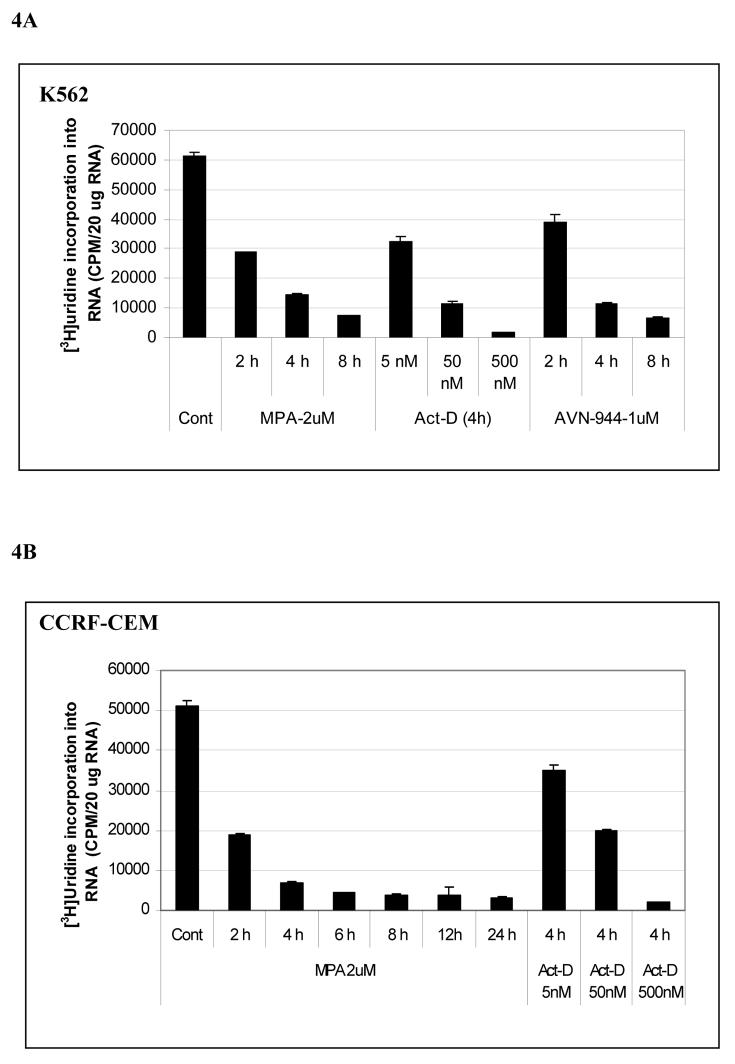
Fig. 4. Effects of MPA, AVN-944, and Actinomycin D on total RNA synthesis and prerRNA synthesis in K562 and CCRF-CEM cells.


K562 cells were treated with MPA, AVN-944, or actinomycin-D at the concentrations and times indicated and [3H]-uridine incorporation into RNA was measured (A, B); the effect of MPA on the expression of pre-rRNA levels was determined by RT-PCR (C) as described in Materials and Methods. Primers amplified the 394 bp fragment extending from the 3' end of 18 S rRNA to the internal transcribed sequence (ITS) located between 18 S rRNA and 5.8 S rRNA of the intact pre-rRNA (shown as model figure, C). Analysis of rRNA processing (D). Extracted total [3H]-uridine-labeled RNAs from untreated K562 cells and cells treated with 2 μM MPA or 1 μM AVN-944 were separated on 1% agarose gel containing formaldehyde, transferred to membrane, and detected by autoradiography as described in Materials and Methods. The bottom panel shows 28S and 18S ribosomal RNA after transfer.