Abstract
Antibody-conjugated nanomaterials have attracted much attention because of their applications in nanomedicine and nanotheranostics, and amplification of detection signals. For many of these applications, the nano-conjugates must bind with a cell membrane receptor (antigen) specifically before entering the cells and reaching the final target, which is thus important but not well understood. Here we present a plasmonic imaging study of the binding kinetics of antibody-conjugated gold nanoparticles with antigen-expressing cells, and compare the results with that of the nanoparticle-free antibody. We find that the nano-conjugates can significantly affect the binding kinetics compared with free antibody molecules, depending on the density of the antibody conjugated on the nanoparticles, and expressing level of the antigen on the cell membrane. The results are analyzed in terms of a transition from mono-valent binding model to a bi-valent binding model when the conjugation density and expressing level increase. These findings help optimize the design of functional nanomaterials for drug delivery and correct interpretation of data obtained with nanoparticle signal amplification.
Keywords: Plasmonic image, nano-medicine, binding kinetics, membrane protein
1. Introduction
Nanomaterials have been used as smart carriers for drug and gene delivery,[1, 2] versatile probes for diagnostic imaging,[3] and potential agents for thermotherapy.[4, 5] For most of these applications, the nanomaterials are functionalized with antibodies to form nano-conjugates so that they can recognize the target cells and cross the cell membranes in order to reach a target location in the cells.[6–8] The recognition is based on the specific binding of the surface-bound antibody with a molecular receptor expressed by the cells, e.g., membrane proteins, on the cell membrane surface. The specific antibody-receptor binding also facilitates the subsequent uptake of nano-conjugates via antibody induced receptor endocytosis.[9] Understanding the binding of nano-conjugates with cell membrane is thus critical for many of the proposed biomedical applications with nanomaterials.[10, 11]
Despite the importance, determining how the nano-conjugates interact with membrane receptors has been challenging because of several reasons.[9] First, the binding often involves multiple mechanisms, including specific binding,[9, 12] and less specific electrostatic[13] and hydrophobic interactions.[14] Second, most techniques cannot easily separate the initial binding and subsequent endocytosis processes. Finally, the antibodies bound on the nanomaterial surfaces may interact with the membrane receptors differently from the corresponding antibodies in free solutions. Presently, the interaction of the nanomaterials with cell membranes is described using terms including uptake efficiency and trans-membrane time.[15, 16] A more detailed kinetic model with measurable binding constants would help to quantify the interaction on the molecular scale.[17, 18] Various techniques have been developed to study the interaction of nano-conjugates with cell membranes. For example, optical imaging and tracking techniques have been used to monitor the process of nano-conjugates entering the cells.[19]
In the present work, we quantify the binding kinetics of antibody-functionalized gold nanoparticle (AuNP) with native antigen-expressing cells, and determine the effect of the nanoparticle on the binding kinetics with a plasmonic imaging method. [20] The method allows us to monitor the binding of nano-conjugates onto single intact cells in real time, from which the binding kinetics is studied quantitatively. We choose monoclonal antibody Herceptin-conjugated gold nanoparticles and human epidermal growth factor receptor 2 (Her2)-expressing cells as a model system. Her2 is a receptor tyrosine kinase overexpressed in various ovarian and breast cancers, and Herceptin is one of the most successful monoclonal antibody drugs on the market for treating Her2 positive cancers.[21, 22] The Herceptin-conjugated nanomaterials have shown to exhibit unique advantages in treating cancers.[23–25]
2. Results and Discussions
2.1. Measuring the binding kinetics of Herceptin@AuNP conjugates with SKBR3 cells
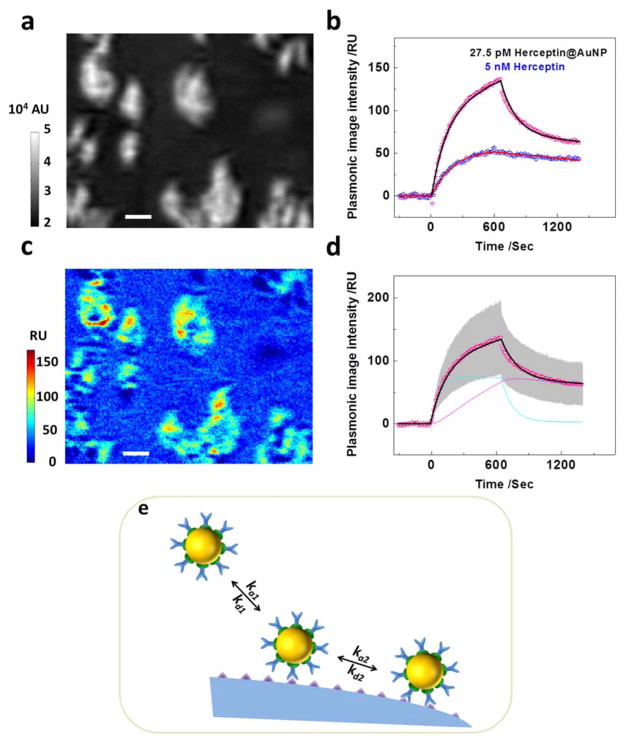
Figure. 1a is the schematic illustration of the plasmonic imaging system for measuring the binding kinetics of nano-conjugates and single adherent cells. Her2-expressing cells were cultured on the sensor chip overnight before the chip was placed on the prism for measurement. Note that the adherent cells were fixed by 4% paraformaldehyde prior to experiment to minimize the intrinsic micromotions of the cells and avoid the possible uptake of nano-conjugates that could affect data interpretation. An incident light was directed through a prism onto the gold-coated coverslip to excite the surface plasmons. A CCD camera was used to capture the reflected light to form a plasmonic image, which maps the distribution of local mass density. Herceptin molecules were immobilized on the AuNP surface with the help of protein A, as shown in Figure. 1b, which bound to Her2-expressing cell membrane via a specific Herceptin-Her2 interaction. The binding of nano-conjugates increased the local mass density of cells, which was measured by the plasmonic imaging. The entire association and dissociation process could be monitored for each single cell, allowing for the extraction of binding kinetics constants at the single cell level.
Figure 1. Monitoring the binding kinetics of nano-conjugates with single intact cells.
(a) Schematic illustration of the plasmonic imaging setup. A p-polarized beam is directed onto the gold-coated glass coverslip through the prism underneath to excite the surface plasmon. The reflected light is captured by a CCD camera to produce a plasmonic image of individual adherent cells. (b) The structure of a Herceptin@AuNP nano-conjugate.
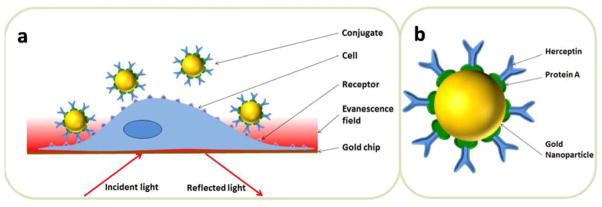
A typical plasmonic image of SKBR3 cells adherent on the gold chip is presented in Figure. 2a, which reveals the individual adherent cells as bright spots. SKBR3 is one of the most widely used breast cancer cell lines that are known to express Her2 endogenously in the cell membranes. The overexpression of Her2 in the cell line was further verified by immunofluorescence staining (Supporting Information Figure. S1). At the beginning of each measurement, buffer solution flowed over the cells for 300 seconds until a stable baseline in the plasmonic image intensity was reached. Then solution containing Herceptin@AuNP was introduced for another 600 seconds to allow the association of nano-conjugates onto the cell membrane. In order to study the dissociation rate constant, the buffer solution was subsequently injected again, flowing over the SKBR3 cells, for 900 seconds. The plasmonic image was continuously recorded throughout the procedure, from which the sensorgrams of the individual cells were obtained. Figure. 2b plots an averaged sensorgram over 36 different cells (black curve), showing well-defined association and dissociation processes.
Figure 2. In situ binding kinetics of nano-conjugates with SKBR3 cells.
(a) A typical plasmonic image of SKBR3 cells, where the bright spots represent individual cells. (b) Typical sensorgrams showing the association and dissociation of nano-conjugates with surface bound Herceptin (black dots) and free Herceptin (blue dots) with SKBR3 cells. The black and red curves are fitted curves with the bi-valent (for pink dots) and mono-valent binding models (for blue dots), respectively. (c) Mapping of nano-conjugate distribution on SKBR3 cells, showing negligible non-specific interactions of nano-conjugates in the gold regions surrounding the cells. (d) Individual sensorgrams of the nano-conjugates with multiple SKBR3 cells (grey), average sensorgram over the individual cells (pink), and the fit of the average sensorgram the bi-valent binding model (black). The cyan curve represents the initial binding of Herceptin to Her2 on the cell membrane via one pair of Herception-Her2 interaction, and the magenta curve is the binding via a second pair of Herception-Her2 interaction. (e) Schematic illustration of the bi-valent binding model.
We further validated that the surface binding of nano-conjugates was indeed from the specific interaction between Herceptin (on the AuNP surface) and Her2 (on the cell membrane). Figure. 2c is a map of mass distribution of the nano-conjugates bound on the individual cells, obtained by subtracting the baseline image before the association (t=0 s) from the one at the end of association (t=600 s). It shows that the nano-conjugates bound on the cells only with little binding onto the surrounding gold surface. As the further controls, we also studied the interactions of Herceptin@AuNPs with Her2-negative cells, and of protein A@AuNP with Her2-positive cells, but observed little binding in both cases (Supporting Information Figure S2). We thus attribute the observed binding of nano-conjugates to the specific Herceptin-Her2 interaction.
In order to evaluate the influence of AuNP on the Herceptin-Her2 binding kinetics, we measured the binding kinetics of free Herceptin molecules with the same batch of SKBR3 cells as shown in Figure. 2b (blue curve). Two features were noticed when comparing the sensorgrams of free (blue curve) and surface-bound Herceptin molecules (black curve). First, the presence of AuNPs amplified the signal by ~two times. This observation is consistent with previous studies and can be understood because AuNPs in general produce a large optical change than the proteins.[26, 27] The second feature was that they followed two different binding kinetics models. While free Herceptin molecules exhibited typical kinetics features of a monovalent model with a dissociation constant (KD) of 2.7 nM (red curve), the nano-conjugates were found to follow a bi-valent model [28, 29] as shown in Figure. 2d (Supporting Information Figure S3). According to this model, Herceptin@AuNP-cell interaction first occurred between one antibody (Herceptin) and one Her2 receptor on the cell membrane. Then an additional molecular binding occurred between another antibody molecule on the same nano-conjugate and an adjacent Her2 receptor to achieve a bivalent attachment.[30] This process is illustrated in Figure. 2e, and a detailed description is provided in Supporting Information.
The bi-valent model is supported by the inter-receptor distance in SKBR3 cells. From the equilibrium response of free Herceptin molecules (54 RU, blue curve), the surface density of Her2 was estimated to be 447 molecules per um2, corresponding to an average inter-molecular distance of 47 nm (Supporting Information Table S1). This distance is much larger than the size of a Herceptin molecule, which explains the observed mono-valent binding for free Herceptin molecules. However, the Herceptin@AuNP conjugate consisted of a 25-nm gold nanoparticle core, a layer of protein A and another layer of Herceptin (Figure. 1b), and its size is comparable to the average inter-molecular distance of Her2, making it possible for two or more Herceptin molecules in the same nano-conjugate to bind to two Her2 receptors on the cell membrane (Supporting Information Figure S4). The formation of one-nanoconjugate-multiple-Her2 therefore resulted in the multi-valency feature as observed in the sensorgrams (Figure. 2d). To confirm this hypothesis, we measured the binding signal of nano-conjugates with 6 nm AuNP and observed mono-valent binding (Supporting Information Figure S5). Such result further demonstrated that the plasmonic imaging technique was reliable for determining the binding kinetics of nano-conjugates with intact cells. Note that even though the experimental results fitted with the bi-valent model very well, multi-valent binding might also exist because of the inhomogeneous distribution of Her2 receptors in the cell membrane.
2.2. Binding kinetics of Herceptin@AuNP with JIMT1
In order to further examine the bi-valent kinetics model, we studied another cell line JIMT1, which was reported to express five times less amount of Her2 than SKBR3.[31] Figure. 3a displays the plasmonic images of several adherent JIMT1 cells. We first obtained the sensorgram of free Herceptin with JIMT1 cells as shown in Figure. 3c (blue curve). The lower expression level of Her2 in JIMT1 was confirmed by the observation of a smaller amount of Herceptin binding (16 RU). We estimated the surface density of Her2 to be ~132 molecules per um2 with an intermolecular distance of 87 nm (Supporting Information Table S1).
Figure 3. In situ binding kinetics of nano-conjugates with JIMT1 cells.
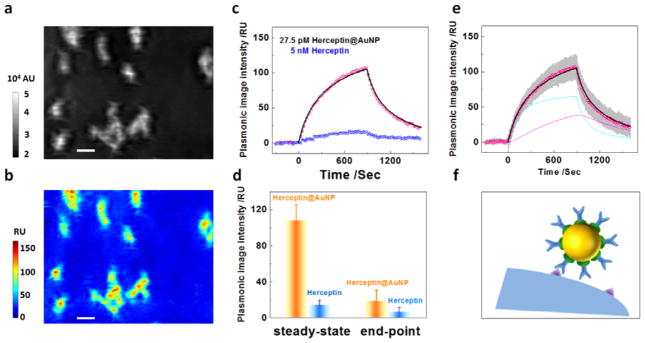
(a) A typical plasmonic image of several JIMT1 cells adherent on a gold chip. (b) Mass distribution of nano-conjugates on JIMT1 cells, showing negligible non-specific interactions of nano-conjugates in the surrounding gold regions. (c) Sensorgrams showing the association and dissociation of nano-conjugates (pink dots) and free Herceptin (blue dots) with JIMT1 cells, where the black curve is a fit with the bi-valent binding model. (d) Plasmonic signal amplification due to the presence of gold nanoparticles. (e) Individual sensorgrams of the nano-conjugates with multiple JIMT1 cells (grey), average sensorgram over the different cells (pink), where the fit of the average sensorgram the bi-valent binding model (black). The cyan curve represents the initial binding of Herceptin to Her2 on the cell membrane via one pair of Herception-Her2 interaction, and the magenta curve is the binding via a second pair of Herception-Her2 interaction. (f) Schematic illustration of the loss of bi-valent binding due to increased inter-molecular distance of Her2 receptors.
We subsequently studied the binding kinetics of nano-conjugates with Her2 receptors on JIMT1 cells (black curve) and generated the binding map of nano-conjugates shown in Fig 3b. Similar to that on SKBR3, the presence of AuNP enhanced the binding signal by ~5 times as displayed in Figure. 3d. However, the intensity dropped rapidly to ~20 RU in 10 minutes during the dissociation stage, implying a weak nano-conjugate-Her2 binding in JIMT1 cells. We also used the bivalent model to fit the sensorgram as shown in Figure. 3e, and found that the monovalent attachment dominated the binding process of nano-conjugates on the cell membrane of JIMT1. This result is consistent with the hypothesis that the small intermolecular distance between Her2 receptors was responsible for the bi-valent binding of nano-conjugates. As the receptor density decreased, the average inter-molecular distance increased, which reduced the probability of bi-valent binding as illustrated in Figure. 3f. Thus, the monovalent attachment became dominant for Herceptin@AuNP-cell interaction with JIMT1 cells.
The binding kinetic constants of Herceptin@AuNP conjugates with SKBR3 and JIMT1, as well as free Herceptin to SKBR3, were also listed in Table 1 for comparison. It is obvious that nano-conjugates exhibited faster association rate constant (ka1) than free Herceptin molecules. However, the dissociation rate constants (kd1) were also much faster for nano-conjugates, resulting in weaker binding affinity than that of free Herceptin molecules. This is probably because the negative charge of nano-conjugate inhibited its accessibility with the cell membrane, which is also negatively charged under physiological conditions. Moreover, nano-conjugates showed much slower dissociation rate constant (kd2) of SKBR3 cells than that of JIMT1 cells, which was due to a higher probability of bi-valent binding of nano-conjugates with Her2-overexpressing SKBR3 cells.
Table 1.
Kinetic rate constants of nano-conjugates and free Herceptin with Her2 receptors in intact SKBR3 and JIMT1 cells
| ka1 (106 M−1s−1) | kd1 (10−3 s−1) | KD1 (nM) | ka2 (10−6 RU·s−1) | kd2 (10−3 s−1) | |
|---|---|---|---|---|---|
| Herceptin@AuNP-JIMT1 | 0.62±0.07 | 8.45±6.15 | 13.32±8.71 | 0.12±0.09 | 1.17±0.64 |
| Herceptin@AuNP-SKBR3 | 2.81±1.64 | 16.64±4.73 | 8.06±4.82 | 0.85±0.36 | 0.26±0.11 |
| Herceptin-SKBR3 | 0.13±0.04 | 0.34±0.26 | 2.70±2.01 |
2.3. Effect of Herceptin density in Herceptin@AuNP conjugates on the binding kinetics
The number of antibodies on a single nanoparticle may also affect the interaction of the nano-conjugates with the cells, and subsequently affect the performance of the nano-conjugates on diagnostics, drug delivery and therapy. To study this effect, we prepared nano-conjugates with different Herceptin to AuNP molar ratios, and studied its influence on the binding kinetics of nano-conjugates with cell membranes on SKBR3 cells. As shown in Figure. 4a the maximum binding amount of nano-conjugates increased with the density of Herceptin conjugated on AuNPs, where the conjugation density was estimated from the molar ratio between Herceptin molecules and gold nanoparticles and all the nano-conjugates were adjusted to be the same concentration of 5.5 pM in the experiments for fair comparisons. A close analysis of the binding kinetics showed that the monovalent binding model fitted the data well when the Herceptin density was relatively low (Figure. 4b). When the molar ratio of Herceptin to AuNP increased from 25 to 150, the sensorgram followed a typical bi-valent model as shown in Figure. 4c (Supporting Information Figure S6). We believe that the increase of Herceptin conjugation density enhanced the binding possibility of multiple Herceptin molecules in one nano-conjugate with Her2, i.e., multi-valent binding. These results indicate that the binding kinetics of nano-conjugates indeed depends on the conjugation density of antibody, which must be considered when designing nano-conjugates for efficient drug or gene delivery with nano-conjugates.
Figure 4. Influence of Herceptin conjugation density on the binding mechanisms of nano-conjugates.
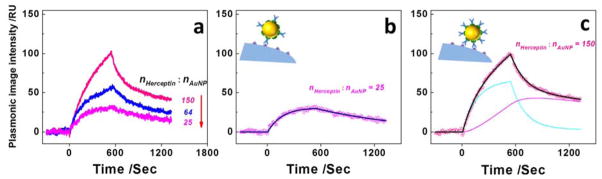
(a) Sensorgrams with Herceptin to AuNP molar ratios at 150 (pink curve), 64 (blue curve) and 25 (magenta curve), respectively. (b) The sensorgram of nano-conjugates with the lowest Herceptin to AuNP molar ratio fits typical monovalent binding model (panel b inset). (c) The sensorgram of nano-conjugates with the highest Herceptin to AuNP molar ratio fits the bi-valent binding model (panel c inset). Cyan and magenta lines represent the initial mono-valent attachment and the formation of bi-valent binding, respectively.
Finally, we point out that when Her2 expressing levels were low, the binding signals of free Herceptin molecules with the cells could not be easily detected, and the presence of AuNPs significantly enhanced the plasmonic signal, which is a useful method to amplify weak binding signals. However, one must optimize the density of Herceptin molecules on the AuNPs and carefully analyze the kinetic data in order to avoid the effect of AuNPs on the binding kinetics.
3. Conclusion
We have studied the binding of Herceptin-conjugated gold nanoparticles with intact Her2-expressing cells using a plasmonic imaging technique, and compared the results with that of free Herceptin the Her2-expressing cells. By analyzing the binding kinetics, we conclude that the nano-conjugates can generally amplify the plasmonic signals, but also significantly affect the binding kinetics. The effect depends on the conjugation density of Herceptin on AuNPs and the expressing level of Her2 on the cell membrane. If the density of Herceptin and/or the expressing level of Her2 are low, the binding kinetics follows a simple mono-valent model. However, if the density of Herceptin and the expressing level of Her2 are high, the binding kinetics can only be described by a bi-valency model, involving two pairs of specific Herceptin-Her2 interactions. The present work demonstrates a method for direct study of the binding kinetics of nano-conjugates with cell membranes, and also provides new insights into the mechanisms underlying the recognition and uptake of nano-conjugates by cells. These results show that one must consider the effects of nanomaterials on the binding kinetics when designing nano-conjugates for efficient drug or gene delivery with nano-conjugates, or using nanoparticles to amplify detection signals.
4. Experimental Section
Materials
Herceptin was obtained in powder form Genetech Inc. (San Francisco, CA), and its stock solution was prepared by dissolving 42 mg powder in 1 mL deionized water, reaching an effective antibody concentration of 21 mg/mL by following the manufacturer’s instruction. AuNP coated with protein-A (AuNP-PrA, AuNP diameter = 25 nm, coefficient of variance: <12%) was purchased from Electron Microscope Sciences (Hatfield, PA). Paraformaldehyde of 4% (v/v) was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Phosphate-buffered saline (1xPBS, pH = 7.4) purchased from Thermo Fisher (Waltham, MA) was used as the buffer solution. The buffer solution was adjusted with deionized water and sodium chloride to reach the same refractive index as the sample solution containing nano-conjugates for studying the binding of nano-conjugates. All other solvents and chemicals were analytical grade purchased from Sigma Aldrich unless stated otherwise.
Preparation of gold nanoparticle–Herceptin conjugates (Herceptin@AuNP)
According to the manufacturer’s instruction, approximately 150 Protein A (PrA) molecules are covalently coated on each AuNP. The conjugation of Herceptin was achieved by further incubating AuNP-PrA with Herceptin as their application protocol. The conjugation density, i.e., Herceptin to AuNP molar ratio, was adjusted by modifying the concentration of Herceptin during conjugation process. Briefly, 100 nM Herceptin was mixed with 0.55 nM AuNP-PrA in 500 μL deionized water and the mixture was left overnight in the dark in order to achieve the highest Herceptin to AuNP molar ratio of ~150. This solution was further diluted with 1xPBS to reach the experimental concentration as indicated in the text. Herceptin@AuNPs with different conjugation densities were prepared in a similar way using different Herceptin concentrations. And nano-conjugates with highest molar ratio of Herceptin to AuNP were used in the present work unless stated otherwise. Such a conjugating protocol allowed PrAs to specifically recognize and oligomerically bind to the Fc domain of Herceptin molecules, so that Herceptin molecules could adapt optimal orientation for targeting Her2 receptors on the cell membrane.[32] Note that the link of PrA with AuNPs and PrA-Herceptin interactions were very stable and tight. There was no desorption or crosslinking of protein from the nanoparticle once the conjugation is formed. [33]
Cell culture
SKBR3 cell line were obtained from the American Type Culture Collection (ATCC, Rockville, MD) and cultured according to their specifications. JIMT1 cell line was purchased from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 10% Fetal Bovine Serum (FBS, Invitrogen) and penicillin-streptomycin (BioWhittaker, Basel, Switzerland) were used as culture media. When the cells reached the confluent of approximately 80%, they were passaged with 0.05% trypsin and 0.02% EDTA in Hank’s balanced salt solution (HBSS, Sigma-Aldrich, St. Louis, MO).
Plasmonic imaging setup
A prism-based plasmonic imaging configuration with 10X magnification was used for collecting the binding kinetics information. The detailed description of this setup can be found in our previous work.[22] Briefly, p-polarized light from a 670 nm light-emitting diode (LED, L7868-01, Hamamatsu, Japan) was directed onto the sensor chip through a SF-11 prism. The reflected beam by the prism produced a plasmonic image, which was captured by a CCD camera (Pike F032B, Allied Vision Technologies, Newburyport, MA).
The sensor chips were BK-7 glass coverslips coated with 2~3 nm chromium followed with ~47 nm gold. A Flexi-Perm silicon chamber (SARSTEDT, Germany) was placed on top of the gold chip to serve as a cell culture well. To promote strong adhesion of cells on the gold surface, the chips were pretreated with 75 μL of 10 μg/mL collagen solution. About 5000 cells in 250 μL growth media were added into the chamber. The wells were kept in the incubator for 24 hours to allow the attachment and growth of cells. Prior to measuring kinetics, the adhered cells were rinsed by PBS buffer, and then fixed by 4% paraformaldehyde solution.
Flow system
A gravity-based multichannel drug perfusion system (SF-77B, Warner Instruments, CT) was used to control the local solution surrounding the target cells. The typical transition time between different solutions was about 1–2 seconds.
Supplementary Material
Acknowledgments
We thank Genentech Inc. for providing the Herceptin used in the present research, and the financial supports from National Natural Science Foundation of China (NSFC, Grant nos. 21327008, 21405080), Natural Science Foundation of Jiangsu Province (BK20140592), and National Institutes of Health (#1R01GM107165-01A1).
Footnotes
Supporting Information as indicated in the text is available from the Wiley Online Library or from the author.
Contributor Information
Linliang Yin, School of Chemistry and Chemical Engineering, Chongqing University, No.174, Shazheng St., Shapingba Dist., Chongqing 400044, China. 1001 S. McAllister Ave. Tempe, AZ 85287, United State.
Yunze Yang, 1001 S. McAllister Ave. Tempe, AZ 85287, United State.
Dr. Shaopeng Wang, 1001 S. McAllister Ave. Tempe, AZ 85287, United State
Prof. Wei Wang, State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, Jiangsu 210093, China
Prof. Shengtao Zhang, Email: stzhang@cqu.edu.cn, School of Chemistry and Chemical Engineering, Chongqing University, No.174, Shazheng St., Shapingba Dist., Chongqing 400044, China
Dr. Nongjian Tao, Email: njtao@asu.edu, 1001 S. McAllister Ave. Tempe, AZ 85287, United State. State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, Jiangsu 210093, China
References
- 1.Yang XJ, Liu X, Liu Z, Pu F, Ren JS, Qu XG. Adv Mater. 2012;24:2890–2895. doi: 10.1002/adma.201104797. [DOI] [PubMed] [Google Scholar]
- 2.Chen CE, Geng J, Pu F, Yang XJ, Ren JS, Qu XG. Angew Chem-Int Edit. 2011;50:882–886. doi: 10.1002/anie.201005471. [DOI] [PubMed] [Google Scholar]
- 3.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 4.Davis ME, Chen Z, Shin DM. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 5.Petros RA, DeSimone JM. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 6.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 7.Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 8.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharyya S, Bhattacharya R, Curley S, McNiven MA, Mukherjee P. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14541–14546. doi: 10.1073/pnas.1006507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu K, Sun J, Bickford LR, Lin AWH, Halas NJ, Yu TK, Drezek RA. Nanotechnology. 2008;19:6. doi: 10.1088/0957-4484/19/04/045103. [DOI] [PubMed] [Google Scholar]
- 11.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Holl MMB. Chem Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Gao HL, Yang Z, Zhang S, Cao SJ, Shen S, Pang ZQ, Jiang XG. Scientific Reports. 2013;3 doi: 10.1038/srep02534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha K, Kim ST, Yan B, Miranda OR, Alfonso FS, Shlosman D, Rotello VM. Small. 2013;9:300–305. doi: 10.1002/smll.201201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam HY, Kwon SM, Chung H, Lee SY, Kwon SH, Jeon H, Kim Y, Park JH, Kim J, Her S, Oh YK, Kwon IC, Kim K, Jeong SY. Journal of Controlled Release. 2009;135:259–267. doi: 10.1016/j.jconrel.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Lesniak A, Salvati A, Santos-Martinez MJ, Radomski MW, Dawson KA, Aberg C. J Am Chem Soc. 2013;135:1438–1444. doi: 10.1021/ja309812z. [DOI] [PubMed] [Google Scholar]
- 16.dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. Small. 2011;7:3341–3349. doi: 10.1002/smll.201101076. [DOI] [PubMed] [Google Scholar]
- 17.Zhang RM, Monsma F. Expert Opinion on Drug Discovery. 2010;5:1023–1029. doi: 10.1517/17460441.2010.520700. [DOI] [PubMed] [Google Scholar]
- 18.Swinney DC. Current Opinion in Drug Discovery & Development. 2009;12:31–39. [PubMed] [Google Scholar]
- 19.Gu Y, Sun W, Wang GF, Fang N. J Am Chem Soc. 2011;133:5720–5723. doi: 10.1021/ja200603x. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Yang YZ, Wang SP, Nagaraj VJ, Liu Q, Wu J, Tao NJ. Nat Chem. 2012;4:846–853. doi: 10.1038/nchem.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams GP, Weiner LM. Nat Biotechnol. 2005;23:1147–1157. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Yin LL, Gonzalez-Malerva L, Wang SP, Yu XB, Eaton S, Zhang ST, Chen HY, LaBaer J, Tao NJ. Sci Rep. 2014;4:7. doi: 10.1038/srep06609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi Y, Liu X, Zhao J, Ding J, Feng SS. Biomaterials. 2012;33:7519–7529. doi: 10.1016/j.biomaterials.2012.06.100. [DOI] [PubMed] [Google Scholar]
- 24.Zhang K, Hao L, Hurst SJ, Mirkin CA. J Am Chem Soc. 2012;134:16488–16491. doi: 10.1021/ja306854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arruebo M, Valladares M, Gonzalez-Fernandez A. Journal of Nanomaterials. 2009 [Google Scholar]
- 26.Zeng S, Baillargeat D, Ho HP, Yong KT. Chemical Society Reviews. 2014;43:3426–3452. doi: 10.1039/c3cs60479a. [DOI] [PubMed] [Google Scholar]
- 27.Lyon LA, Musick MD, Natan MJ. Anal Chem. 1998;70:5177–5183. doi: 10.1021/ac9809940. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins JL, Lee MK, Valaitis AP, Curtiss A, Dean DH. J Biol Chem. 2000;275:14423–14431. doi: 10.1074/jbc.275.19.14423. [DOI] [PubMed] [Google Scholar]
- 29.Yang TL, Baryshnikova OK, Mao HB, Holden MA, Cremer PS. J Am Chem Soc. 2003;125:4779–4784. doi: 10.1021/ja029469f. [DOI] [PubMed] [Google Scholar]
- 30.Pisarchick ML, Thompson NL. Biophys J. 1990;58:1235–1249. doi: 10.1016/S0006-3495(90)82464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy P, Friedlander E, Tanner M, Kapanen AI, Carraway KL, Isola J, Jovin TM. Cancer Res. 2005;65:473–482. [PubMed] [Google Scholar]
- 32.Atkins KL, Burman JD, Chamberlain ES, Cooper JE, Poutrel B, Bagby S, Jenkins ATA, Feil EJ, van den Elsen JMH. Mol Immunol. 2008;45:1600–1611. doi: 10.1016/j.molimm.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Zrazhevskiy P, Gao XH. Nat Commun. 2013;4:12. doi: 10.1038/ncomms2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.