Figure 7.
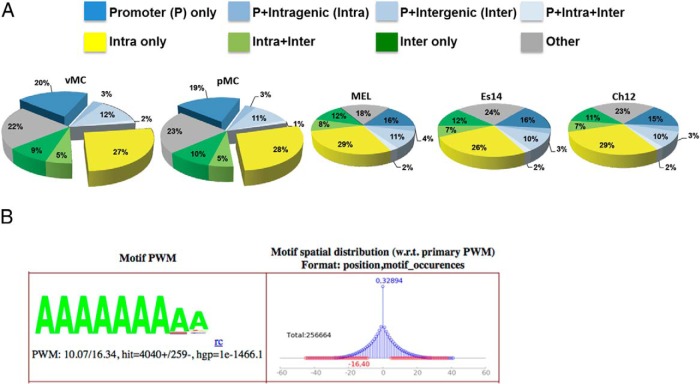
Nmp4 binds to AT-rich DNA typically proximal to TSS sites or within intragenic regions. A, Genome-wide mapping of the Nmp4 binding sites show that most sites are distributed in the TSS and intragenic regions of the genome. ChIP-seq analysis included vehicle-treated and PTH-treated MC3T3-E1 osteoblast-like cells (vMC and pMC, respectively) and 3 murine cell lines from the ENCODE Consortium, including ES-E14 (Es14), which are E14 undifferentiated mouse embryonic stem cells, and 2 mouse erythroleukemia cell lines (Ch12 and MEL) derived from B cell lymphomas. B, GEM analysis for the Nmp4 consensus sequence derived from MC3T3-E1 cells. A minimal k-mer width of 6 and maximum of 20 was used. The optimal position weight matrix (PWM) score for the MC3T3-E1 data was 10.07. The hypergeometric P value (hgp) was 1e-1466.1.