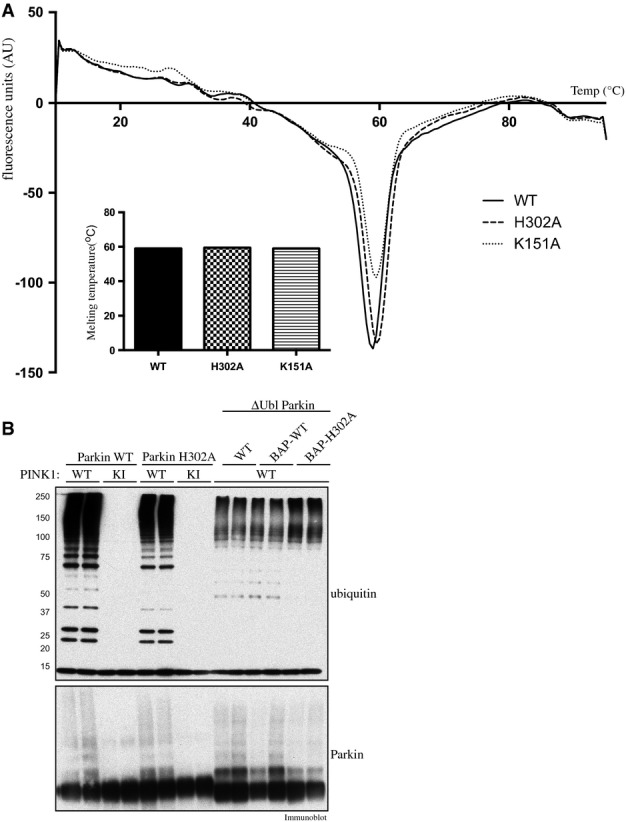
Analysis of stability and ubiquitinPhospho-Ser65-independent E3 ligase activity of “Pocket 2” mutants of Parkin
- Thermal denaturation curves obtained by differential scanning fluorimetry of wild-type (WT), and His302Ala (H302A)- and Lys151Ala (K151A)-mutant Parkin. Results are displayed as the differential of the fluorescence in arbitrary units divided by the differential of the temperature, plotted against temperature. Inset: the minimum of each curve indicates the melting point (Tm). Summary of melting points (Tm) of each protein as follows: WT (59°C); H302A (59.5°C); and K151A (59.5°C).
- PINK1-dependent full-length Parkin E3 ligase activity mediated via phosphorylation of Ubl Ser65 and constitutive basal E3 ligase activity mediated by Ubl-deleted Parkin (ΔUbl; residues 80-465) are not affected by His302Ala (H302A) mutation. A 2 μg amount of wild-type full-length or ΔUbl-Parkin-biotin and H302A full-length or ΔUbl-Parkin was incubated with 1 μg of wild-type (WT), kinase-inactive (KI) or no TcPINK1 in an E3 ligase assay. Reactions were terminated after 60 min by the addition of LDS loading buffer and analysed by SDS/PAGE. Ubiquitin and Parkin were detected using anti-FLAG and anti-Parkin antibodies, respectively.
