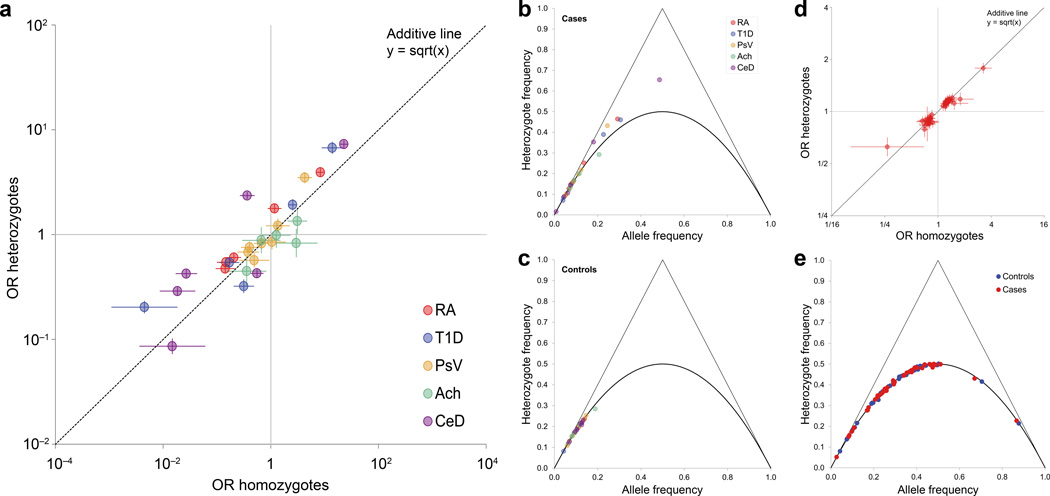
Figure 1. Disease associations of HLA and non-HLA variants.
(a) Disease associations of HLA haplotypes with rheumatoid arthritis (RA), type 1 diabetes (T1D), psoriasis vulgaris (PsV), idiopathic achalasia (Ach), and celiac disease (CeD). For each common haplotype, the odds ratio (OR) for heterozygotes (vs. non-carriers) is plotted against the OR for homozygotes (vs. non-carriers). The dashed line represents a purely log-additive relationship, in which heterozygotes have exactly half the risk of homozygotes (on a log-odds scale). Data points above the dashed line represent haplotypes with a positive dominance component, and below the line haplotypes with a negative dominance component. Error bars represent 95% confidence intervals. (b,c) De Finetti diagram of the proportion of heterozygotes in relation to the frequency of each HLA haplotype (grouped across all diseases), shown separately for (b) cases and (c) controls. The solid line represents the expected proportion of heterozygotes under Hardy-Weinberg-Equilibrium. (d) Disease association of 43 known genome-wide RA-associated SNPs located outside the MHC region, using the same plotting scheme as for panel a. No single SNP shows a significant deviation from the dashed line (representing a purely additive disease contribution). (e) De Finetti diagram of heterozygote frequency for the same 43 non-MHC SNPs as in panel d, given separately for controls and cases.