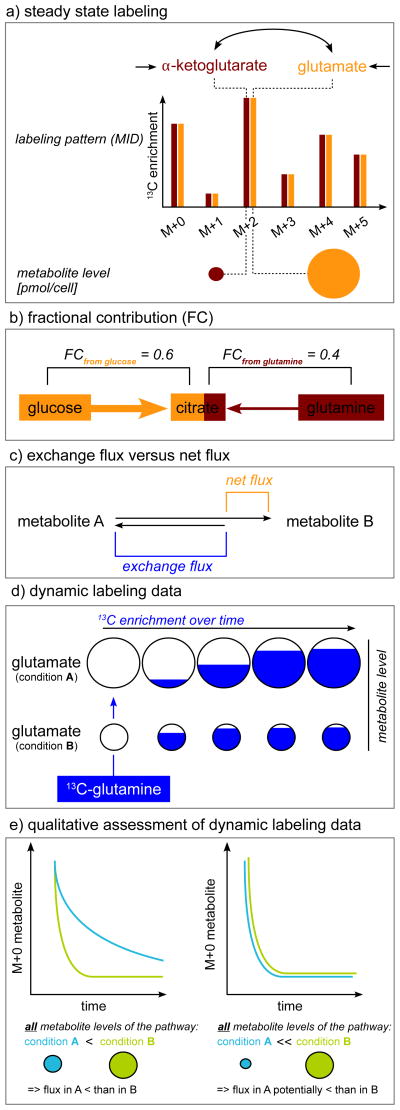
Figure 2.
Interpretation of labeling data. a) Steady state labeling data are independent from the metabolite levels. b) Fractional contribution quantifies the contribution of a labeled nutrient to the metabolite of interest. c) Exchange fluxes can lead to rapidly labeled metabolites although the net flux of the nutrient to the metabolites is small. d) Dynamic labeling patterns are metabolite level dependent: The flux from glutamine to glutamate is the same in condition A and B, but in condition A the glutamate levels are greater than in condition B. Consequently, the labeling dynamics of glutamate in condition A are slower than in condition B although the flux from glutamine to glutamate is the same in both conditions. e) Relative flux activity between two conditions can be evaluated without kinetic flux calculations if both the labeling dynamics and all metabolite levels of the pathway of interest are altered in the same direction.