Abstract
Cutaneous T cell lymphoma (CTCL) is a non-Hodgkin lymphoma of skin-homing T lymphocytes. We performed exome and whole genome DNA sequence and RNA sequencing on purified CTCL and matched normal cells. The results implicate mutations in 17 genes in CTCL pathogenesis, including genes involved in T cell activation and apoptosis, NFκB signaling, chromatin remodeling, and DNA damage response. CTCL is distinctive in that somatic copy number variants (SCNVs) comprise 92% of all driver mutations (mean of 11.8 pathogenic SCNVs vs. 1.0 somatic single nucleotide variants per CTCL). These findings have implications for novel therapeutics.
Introduction
Cutaneous T cell lymphoma (CTCL) is a malignancy of skin-homing T cells. Patients typically present with localized patches and plaques in sun-protected skin. Lymphoma cells may spread from these lesions to uninvolved skin and proliferate in the dermis, leading to thicker plaques and tumors. In advanced disease, malignant T cells disseminate to blood, lymph nodes, and viscera. In leukemic CTCL,1 malignant T cells can comprise >99% of circulating T lymphocytes. Loss of the normal T cell receptor (TCR) repertoire leads to immunosuppression and opportunistic infections, which are the most common disease-related causes of death.
CTCL cells display constitutive activation of the T cell receptor pathway1, constitutively produce TCR-dependent Th2 cytokines such as IL-4 and IL-132 and are resistant to normal mechanisms that prevent uncontrolled proliferation, including FAS-mediated apoptosis and growth-suppression via TGF-β3.
There has been limited genome-level study of mutations in CTCL4 and there are no genetic biomarkers guiding diagnosis and treatment5. Recurrent deletions of 10q and 17p and amplifications of 8q and 17q have been identified, with robust evidence implicating deletions of TP53 and CDKN2A, and amplification of 8q containing MYC5–11. However, there has been little consensus regarding the target genes underlying other recurrent copy number mutations12. Similarly, candidate gene sequencing has suggested a role for a recurrent gain-of-function point mutation in PLCG111. However, reports of genome level sequencing has thus far been limited to a single sample. We report herein the genomic landscape of CTCL from the study of 40 cases.
Results
Purification of CTCL cells and next generation sequencing
We studied 40 patients with stage IVA1-B disease and CTCL cells in blood. The mean time from diagnosis to sampling was 2.5 years. 37 patients had been treated with extracorporeal photopheresis (ECP). We performed flow cytometry using highly specific cell surface markers to purify CTCL cells and matched normal monocytes (Methods and Supplementary Fig. 1).
The resulting CTCL cells and normal monocytes had a median purity >97% and were subjected to exome sequencing (see Methods); the median coverage was 220 and 184 independent reads per targeted base, respectively. For 11 CTCLs, we also performed RNA-sequencing with a median of 43.5 million 74 bp paired-end reads per sample. Two of these also had whole genome sequencing (WGS) (mean coverage depth of 33 independent reads per targeted base (see Methods and Supplementary Table 1)).
Somatic single nucleotide variants (SSNVs) were called from comparison of the read distributions in CTCLs and matched normal exomes (see Methods), identifying a median of 63 SSNVs per sample (range 13 to 175) (Fig. 1a). 74% of the mutations were C>T transitions, predominantly occurring in a dipyrimidine context, consistent with induction by ultraviolet light13. CTCLs with and without ECP showed similar proportions of these mutations (Supplementary Table 2).
Figure 1. Landscape of somatic alterations in CTCL.
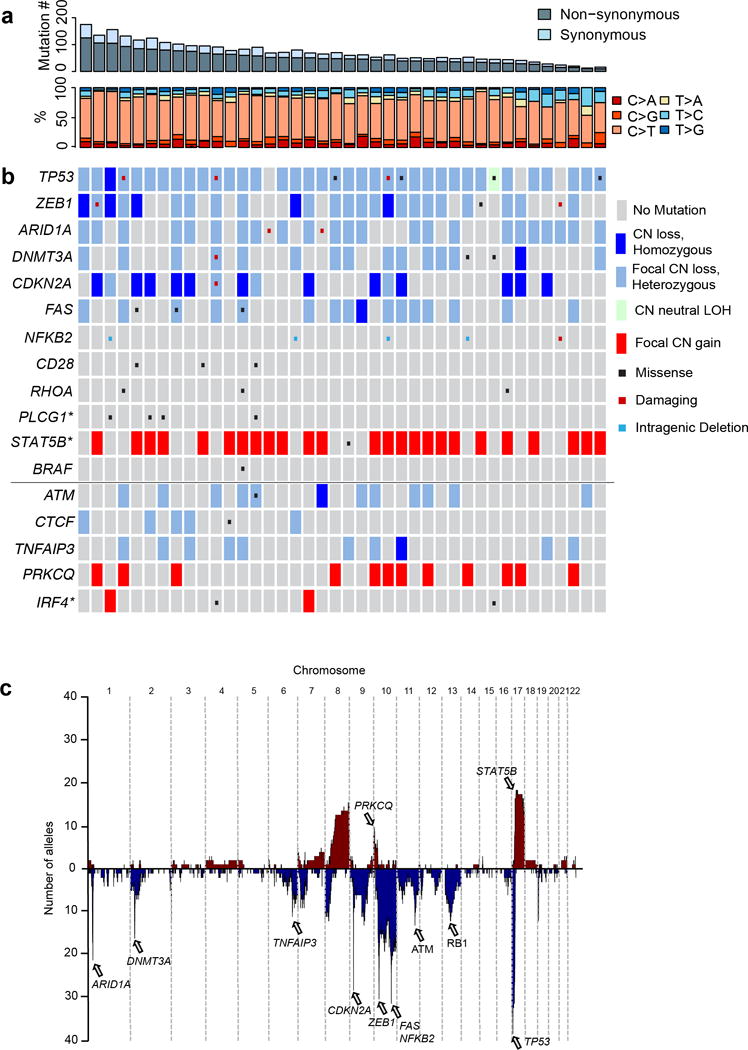
Each column shows data for one CTCL. (a) The number of SSNVs in each CTCL along with the relative frequency of transitions and transversions is shown. (b) Select significant somatic alterations identified by whole exome sequencing are shown. Genes above the solid horizontal line have significant support for gene localizing mutations. Genes shown below the solid horizontal line represent putative driver genes residing on significant narrow focal SCNVs (< 10 genes in GISTIC interval). * indicates a Q-value between 0.1 and 0.25 supporting the specific gene indicated. (c) Composite plot of somatic copy number abnormalities. Listed are select implicated driver genes.
Somatic copy number variants (SCNVs) were identified by comparing read coverage and minor allele frequencies of heterozygous variants across chromosome segments in CTCL and matched normal exomes (see Methods and Supplementary Fig. 2). CTCLs harbored a mean of 27 SCNVs (two chromosomal arm deletions, two arm amplifications, 17 focal deletions (median size 1.6 Mb), and 6 focal amplifications (median size 2.9 Mb).
99% and 98% of SSNVs and SCNVs respectively called by exome sequencing were also called by WGS (Supplementary Tables 3,4). Selected SSNVs and chromosomal breakpoints were also confirmed by PCR amplification followed by Sanger sequencing (Supplementary Fig. 3, Supplementary Table 5). These data establish high confidence in somatic SSNV and SCNV calls across the cohort.
Somatic Mutations Contributing to CTCL
We implicated specific genes in CTCL pathogenesis using the following approaches (see Methods):
Recurrent SSNVs altering the same amino acid more often than expected by chance (Table 1). This implicated CD28, RHOA and PLCG1.
SSNVs previously identified as recurrent mutations in other cancers (Supplementary Table 6). This implicated BRAF and STAT5B.
Significantly increased burden of protein-altering SSNVs in specific genes; this implicated TP53, DNMT3A, and FAS. (Table 1).
We used GISTIC 2.014 to identify SCNVs that occurred more often than expected by chance, using a Q-value threshold of 0.2515 (Supplementary Tables 7, 8). This identified 29 significant recurrent focal deletions (23 occurring in ≥20% of CTCL, 15 with residual Q-values <0.005) and 7 significant focal amplifications (4 in >20% of CTCL). There were also 4 significant broad deletions and 8 broad amplifications (Supplementary Tables 9, 10). Each CTCL had a mean of 7.5 significant focal deletions (91% heterozygous, 9% homozygous), 1 focal amplification, 1.6 broad deletions, and 1.8 broad amplifications. (Supplementary Fig. 4, Supplementary Tables 7–10). Similar to previously utilized strategies16, we used gene localization data from focal SCNVs and SSNVs to implicate specific genes in seven of these SCNVs (likelihood ratio favoring one gene over all others by >1,000:1; range 2.4 × 103:1 to 6 × 1023:1) (Supplementary Fig. 5, Table 2). Four of the genes implicated are among the pan-cancer set of 127 driver genes17, a result highly unlikely to occur by chance (P= 6 × 10−8; binomial distribution). The results again implicated TP53, DNMT3A, and FAS, and also implicated NFKB2, ARID1A, ZEB1, and CDKN2A.
Table 1.
Genes with significant burden of SSNVs
| Gene | Total SSNVs | Q-value | Recurrent Mutations | Recurrent P-value | SSNVs |
|---|---|---|---|---|---|
| TP53 | 7 | 7.3 × 10−9 | – | – | p.S34X, p.S94X, p.T155N, p.R196X, p.A215V, p.I254T, p.R273P |
| CD28 | 4 | 3.3 × 10−4 | p.F51V, p.Q77P | 9.0 × 10−9 | p.F51I, p.F51V, p.Q77P, p.K81N |
| RHOA | 3 | 0.02 | p.N117I | 1.1 × 10−4 | p.R70K, p.N117I, p.N117I |
| DNMT3A | 4 | 0.06 | – | – | p.P233L, p.Y584X, p.S669F, p.P777L |
| FAS | 3 | 0.07 | – | – | p.S212C, p.E261K, p.D265E |
| PLCG1* | 4 | 0.14 | p.S345F** | 0.006 | p.R48W, p.D342N, p.S345F, p.E1163K |
Gene has a Q-value or false discovery rate between 0.1 and 0.25.
Same mutation found to be a recurrent, gain-of-function mutation in a prior report11
Table 2.
Genes with significant combined burden of somatic CNVs and somatic SNVs.
| Gene | SCNV Type | % of CTCL With SCNV or SSNV | % of CTCL with Deletions | SSNVs | Q-value | Likelihood Ratio | ||
|---|---|---|---|---|---|---|---|---|
| Biallelic | Monoallelic | |||||||
| Focal | Broad | |||||||
| TP53 | Deletion | 92.5 | 2.5 | 7.5 | 80 | NS (3), MS (4) | 5.1 × 10−7 | 1.9 × 106 |
| ZEB1 | Deletion | 65 | 12.5 | 20 | 27.5 | NS (2), MS (1) | 2.8 × 10−19 | 5.9 × 105 |
| ARID1A | Deletion | 62.5 | 0 | 57.5 | 0 | NS (1), FS (1) | 5.7 × 10−24 | 2.4 × 103* |
| DNMT3A | Deletion | 42.5 | 2.5 | 27.5 | 7.5 | NS (1), MS (3) | 2.3 × 10−13 | 2.3 × 105 |
| FAS | Deletion | 42.5 | 2.5 | 10 | 27.5 | MS (3) | 2.2 × 10−3 | 1.3 ×104 |
| CDKN2A | Deletion | 40 | 30 | 7.5 | 2.5 | NS (1) | 3.1 × 10−34 | 6.5 × 105 |
|
| ||||||||
| Gene | SCNV Type | % of CTCL With SCNV or SSNV | % of CTCLs with SCNVs | SSNVs | Q-value | Likelihood Ratio | ||
| Focal | Broad | |||||||
|
| ||||||||
| NFKB2** | C-terminal truncations | 12.5 | 10 | 0 | SS (1) | 5.1 × 10−43 | 5.8 × 1023 | |
NS, nonsense (premature termination) mutation; MS, missense mutation; FS, frameshift mutation; SS, Splice-site mutations. Numbers in parentheses indicate number of CTCLs with SSNVs. Q-value, family-wise error rate for burden of SSNVs and focal SCNVs. Likelihood ratio represents the probability of the total number of somatic mutations in the listed gene occurring by chance divided by the probability of the next most likely gene in the focal SCNV interval.
Likelihood ratio based on number of focal deletions and damaging SSNVs (frameshift, nonsense or splice site mutations).
includes two truncating mutations (one deletion and a splice-site mutation) upstream of exon 17 and three truncating deletions upstream of exon 18.
Collectively, these results identified 12 significantly mutated genes at Q-value threshold of 0.25 (Tables 1,2, Figure 1). Using a Q-value cutoff of 0.1 would eliminate none of these genes; PLCG1 (Q-value > 0.1) is still implicated via a recurrent activating mutation in the PLCx domain11 and supported by clustering of other mutations near the PLCx domain. Mutations in these 12 genes were highly clonal, with 89% of driver mutations found in >75% of tumor cells (Supplementary Table 11).
Additional recurrent SCNVs
We sought additional CTCL drivers among the remaining significant focal SCNVs (see Methods, Supplementary Tables 12, 13). Among seven narrow GISTIC segments containing 9 or fewer genes (mean of 4 genes) we found TNFAIP3 an NF-κB inhibitor recently implicated in CTCL10 (deleted in 25%; only 2 genes in GISTIC interval), We annotated remaining SCNVs for consensus cancer drivers17, which identified ATM (deleted in 30%; 5 genes in GISTIC interval) and CTCF (deleted in 15%; 2 genes in GISTIC interval); finding 2 known cancer drivers among the 28 genes in these seven focal intervals was not expected by chance (P = 0.01; binomial distribution) (Supplementary Tables 14a).
In the remaining four intervals, we used GRAIL18 to identify genes significantly related to other mutated genes, as has been done previously19. PRKCQ, an enzyme in the TCR signaling pathway20 (amplified in 30%; 9 genes in GISTIC interval; GRAIL P-value=6.8 × 10−5) and IRF4, a transcription factor required for expansion of TCR-activated T cells21 (amplified in 5%; 1 of 5 genes in GISTIC interval; GRAIL P-value=0.0015) were implicated (Supplementary Tables 14b). No genes reached significance in the remaining two narrow deletions (Supplementary Table 7, 13).
Collectively, these results implicate 17 genes in CTCL (Figure 1). None showed significant co-occurrence or mutual exclusivity.
Recurrent protein-altering mutations
There were recurrent mutations in six genes, including previously unreported recurrent SSNVs in CD28, and known recurrent mutations in other cancers in RHOA22–24, BRAF25, and STAT5B26, and previously described mutations in two genes previously suggested to play a role in CTCL (PLCG111 and NFKB29) (Tables 1, 2, Fig. 2, Supplementary Tables 5–8).
Figure 2. Recurrent mutations in CTCL.
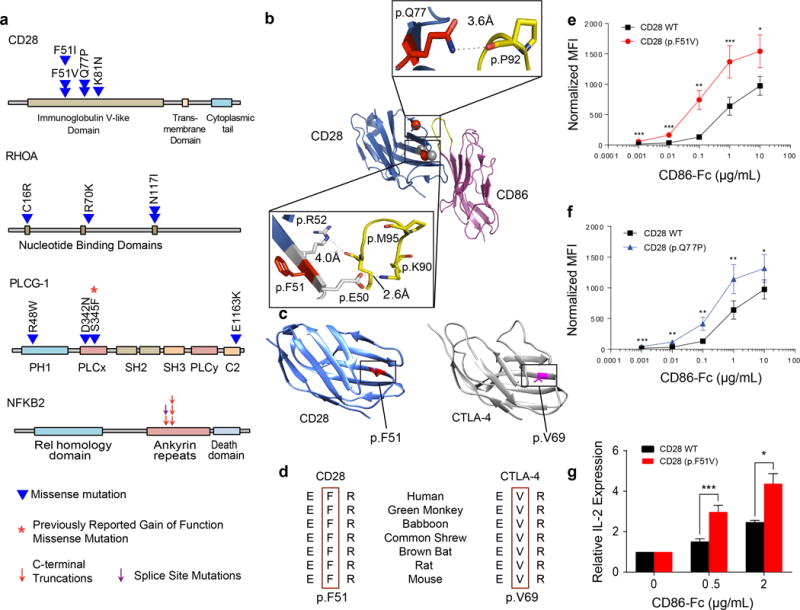
(a) Schematic of SSNVs in CD28, RHOA, PLCG1, and NFKB2. (b) Structural model of CD28-CD86 complex highlighting p.Phe51 and p.Gln77 (red spheres). CD28 coordinates (PDB ID: 1YJD; dark blue) were superimposed on the CTLA-4-CD86 complex (magenta; PDB ID: 1I85). Insets show electrostatic interactions at the indicated amino acids. (c) Locations of p.Phe51 on CD28 (PDB ID: 1YJD; blue) and p.Val69 on CTLA-4 (PDB ID: 1I8L; grey) crystal structures. p.Phe51 and p.Val69 were highlighted red. (d) p.Phe51 on CD28 and p.Val69 on CTLA-4 are highly conserved. E, F, R, and V denote glutamate, phenylalanine, arginine, and valine, respectively. (e–f) Binding of CD86-Fc to 293T cells stably transduced with human wild-type (WT) CD28, (e) CD28 (p.Phe51Val), or (f) CD28 (p.Gln77Pro). CD86-Fc binding was detected by a fluorescently labeled antibody. Fluorescence was then normalized based on the relative surface expression of the CD28 isoform for each sample. Normalized mean fluorescence intensity (MFI) with standard error of seven independent experiments were plotted as a function of CD86-Fc concentration. (g) IL-2 production from Jurkat cells transduced with WT CD28 or CD28 (p.Phe51Val). Jurkat cells were stimulated with phorbol myristate acetate (50 ng/ml), ionomycin (1 μg/ml), and the indicated concentrations of CD86-Fc for 6 hours. Relative IL-2 transcript levels were determined by quantitative RT-PCR, using GAPDH as a control. Graphs plot the means and standard error of 5 biological replicates. Statistical significance was assessed for by two-sided paired ratio t-test (e–g).* P<0.05. ** P<0.005. *** P<0.0005.
CD28 encodes a co-stimulatory T-cell surface molecule that binds B7 ligands (CD80 and CD86) on antigen presenting cells, promoting production of pro-proliferative cytokines and anti-apoptotic Bcl family members27. CD28 harbors four SSNVs (Q= 3.3 × 10−4), all in the extracellular domain. Targeted sequencing of 8 additional CTCLs identified two additional SSNVs, collectively identifying three SSNVs at p.Phe51 and two at p.Gln77, each unlikely to occur by chance (pointwise P-values of P = 9 × 10−9 and 1.3 × 10−4, respectively). The p.Phe51Val mutation was also previously reported in a single case of angioimmunoblastic T cell lymphoma24. Interestingly, these positions are spatially clustered and inferred to be involved in binding of B7 ligands (Fig. 2a,b).
CD28 and CTLA-4 B7 ligand binding domains are homologous, but have opposing effects on T-cell activation28. CTLA-4 and CD28 differ in having valine vs. phenylalaning at homologous positions (p.Val69 in CTLA-4, p.Phe51 in CD28); with flanking amino acids interact with B7 ligands27 (Fig. 2c,d).27 Since CTLA-4 binds to B7 ligands with 50–200-fold greater affinity than CD2827, we hypothesized that the CD28 p.Phe51Val/Ile SSNVs might increase avidity for ligands.
To test this, we used a well-characterized assay29. We expressed wild-type (WT) or mutant CD28 in 293T cells and incubated them with B7 fusion proteins comprising the extracellular domain of B7 fused to human Fc domains. B7 binding was detected with a fluorescently labeled antibody recognizing human Fc domain and binding was adjusted for the level of cell surface CD28. While wild-type and mutant (p.Phe51Val and p.Gln77Pro) CD28s showed no significant difference in binding to CD80-Fc (Supplementary Fig. 6), both mutants demonstrated > 2 – fold higher avidity for binding to CD86-Fc (Fig. 2e,f, Supplementary Fig. 7a,b).
We anticipated that increased avidity of mutant CD28 would cause increased ligand-dependent signaling. We expressed CD28 WT and CD28 p.Phe51Val in Jurkat T cells. Mutant CD28 again showed higher avidity for CD86-Fc (Supplementary Fig. 7 c–f). We compared the ability of WT and mutant CD28 to augment IL-2 expression in response to stimulation with phorbol ester and ionomycin30 in the presence of increasing concentrations of CD86-Fc. Induction of IL-2 was significantly greater with mutant CD28 (Fig. 2g).
RHOA encodes a small GTPase, recently found to be mutated in angioimmunoblastic T cell lymphoma (AITL)23,24, peripheral T cell lymphomas-not otherwise specified (PTCL-NOS)22, and diffuse-type gastric cancer31,32. A recurrent p.Gly17Val SSNV has been shown to be dominant negative22. We found a different recurrent somatic mutation in two CTCLs samples (p.Asn117Ile) (pointwise P = 1.1 × 10−4) (Fig. 2a). Targeted sequencing of 8 more CTCLs revealed an additional p.Cys16Arg SSNV previously seen in PTCL22–24. p.Asn117Ile, p.Cys16Arg, and p.Gly17Val all co-localize in the GTP – binding pocket in the structure of RHOA (Supplementary Fig. 8a). Importantly, both p.Asn117Ile33 and p.Gly17Val22–24 abolish RhoA’s binding of GTP and are dominant negative owing to sequestration of guanine nucleotide exchange factors, impairing formation of GTP-bound molecules24,34.
We also found novel mutations in two oncogenes previously identified in CTCL, PLCG111 and NFKB29. Two PLCG1 mutations (p.Ser345Phe and p.Ser520Phe) which significantly increased enzymatic activity, have been reported11. We found one instance of the p.Ser345Phe mutation and three previously unreported SSNVs, including the nearby p.Asp342Asn, p.Arg48Trp, and p.Glu1163Lys (Figs. 2a, Supplementary Fig. 8b). The recurrence of a known gain of function mutation is unlikely to be a chance event (P = 0.006; binomial distribution), and along with another tightly linked mutation, supports the significance of PLCG1 mutations in CTCL.
We identified 4 deletions with a breakpoint within and one novel splice site mutation in NFKB2, which mediates the non-canonical NF-B pathway. These mutations are tightly clustered and all are inferred to delete the autoinhibitory C-terminus of NFKB2. Similar truncations have been found in lymphomas and result in constitutive activation of the non-canonical NF-κB pathway9 (Table 2, Fig. 2a, Supplementary Fig. 9). In addition, heterozygous deletion of NFKB2 occurred in 57.5% of CTCLs (Supplementary Table 7). This included 13 focal deletions with a minimum common region of only 10 genes. Full-length NFKB2 can inhibit the canonical NF-κB pathway via its C-terminal domain, and its deletion can augment TCR-dependent activation of the canonical NF-κB pathway35.
Lastly, we found known gain-of-function mutations previously found in other cancers. p.Asp594Asn in BRAF is a recurrent oncogenic mutation in melanoma that activates the MAPK pathway25; p.Asn642His in STAT5B is a recurrent oncogenic mutation in CD8+ large granular T cell leukemia26, which increases transcription of STAT5B target genes. Dysregulated STAT5 signaling is critical for CTCL proliferation36 and CTCL’s immunosuppressive effects on normal T cells37. Both BRAF and STAT5B reside within significant focal amplifications found in 10% CTCLs, and when combined with arm-level amplifications are amplified in 17.5% and 62.5% of CTCLs, respectively (Fig. 1b, Supplementary Table 8). Because the GISTIC intervals on 7q34 and 17q11.2 are large, we cannot exclude the possibility of additional target genes on these amplicons. Nonetheless, STAT5B is known to impart oncogenic effects by gene locus amplification alone38,39.
Recurrently deleted genes
The 7 GISTIC intervals with localizing data implicating a single gene included the truncating NFKB2 mutations (discussed above). In addition, six segments that were deleted in 40–90% of CTCLs harbor putative tumor suppressors, including TP53 (SCNVs or SSNVs in 92.5% of CTCLs), which was already implicated by a significant burden of SSNVs. TP53 harbored 4 focal deletions, the minimum overlapping interval of which included TP53 and only 3 other genes, and was also deleted by 32 chromosome arm-level SCNVs. While it is possible that arm-level deletions impart effects via genes in addition to TP53, no other gene on 17p had a statistically significant burden of gene-localizing mutations (Q-values for all other 17p genes = 1). Other known tumor suppressors include CDKN2A (mutated in 40% of CTCLs) and FAS (42.5% of CTCLs). In addition, we found recurrent loss of function mutations in ARID1A (deleted or mutated in 62.5%), DNMT3A (42.5%), and ZEB1 (65%), none of which have previously been implicated in CTCL (Table 2, Figs. 3a–c, Supplementary Figs. 10,11). Mutations in five of these genes were frequently biallelic (Table 2, Fig. 1b), while ARID1A had exclusively heterozygous deletions or damaging SSNVs, consistent with findings in other cancer types40.
Figure 3. Localizing mutations and RNA transcript levels in CTCL.
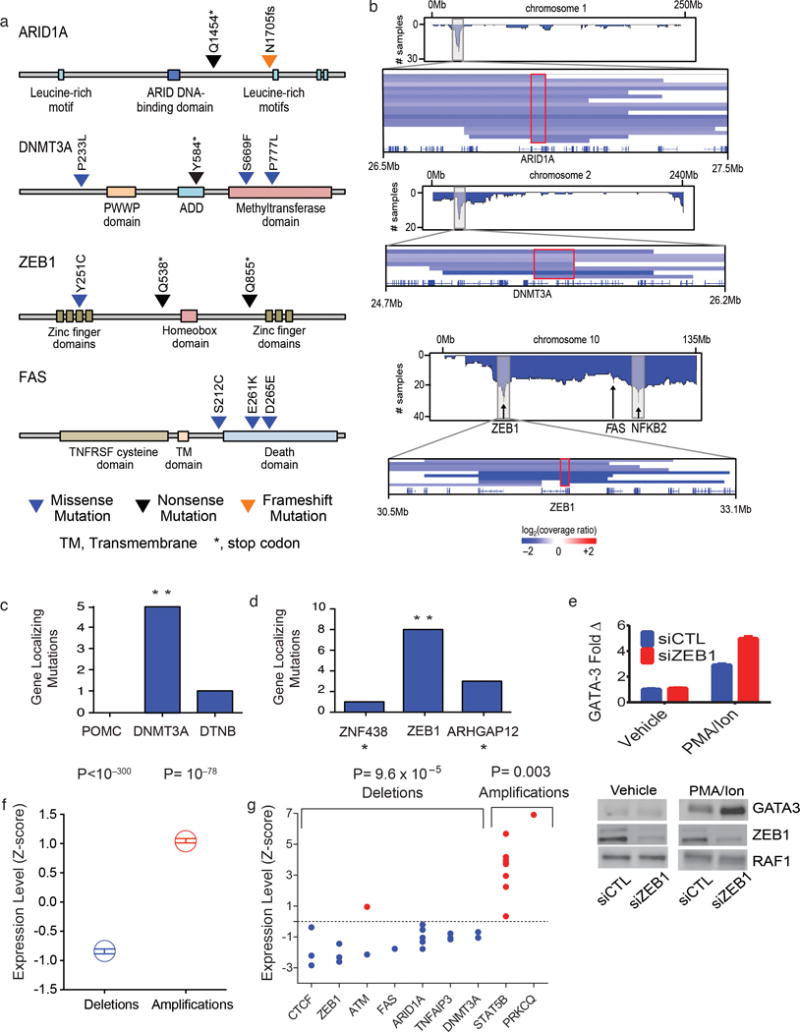
(a) Schematics of SSNVs in ARID1A, DNMT3A, FAS, and ZEB1. (b) Composite plots of deletions across chromosome 1, 2, and 10 with a magnified view of SCNVs at 1p36.11, 2p23.3, and 10p11.22, respectively. Red boxes define minimal common regions (MCR) shared by all SCNVs at each locus. (c–d) Histogram of gene-localizing mutations within the GISTIC confidence interval at 2p23.3 and 10p11.22. Gene-localizing mutations include SSNVs and focal SCNVs that include at least one gene but not all of the genes in the interval. ** likelihood ratio > 1,000:1 supporting one gene as the driver gene in the interval. * genes immediately outside the GISTIC intervals. (e) Increased levels of GATA-3 with ZEB1 knockdown in PMA and Ionomycin (PMA/Ion) stimulated Jurkat cells. Representative experiment of 3 biological replicates is shown. (f) Expression of genes is altered by deletion and amplification. The transcript levels (fragments per kilobase per million reads) of all genes when diploid was normalized to a Z-score of zero with a standard deviation of 1. Mean and standard errors for the transcript levels of the same genes when heterozygously deleted or amplified are shown. (g) Z-scores of putative driver genes with at least 3 CTCLs diploid for the indicated gene. For (f) and (g), statistical significance was calculated using the chi-square test testing the null hypothesis that deleted or amplified genes’ transcript levels are equally likely to be higher or lower than the mean transcript level when the gene is diploid.
ZEB1 encodes a zinc finger transcriptional repressor. ZEB1 is a known oncogene in epithelial cancers41. In contrast, in CTCL as well as in adult T cell leukemia/ lymphoma (ATLL)42, recurrent deletions implicate ZEB1 as a tumor suppressor. In CTCL, six samples harbor biallelic mutations. These include 5 homozygous deletions (range: 1–8 genes homozygously deleted, all including ZEB1) and a deletion in trans to a damaging SSNV (Fig. 1b, Figs. 3a,b). Consistent with this inference, ZEB1 binds to promoters of multiple genes implicated in T cell proliferation and differentiation. For example, decreased ZEB1 expression augments IL-2 transcription43.
Interestingly, ZEB1 binding sites occur upstream of the transcription initiation site of GATA-3, a transcription factor whose expression is sufficient for Th2 T-cell polarization44. GATA-3 is overexpressed in TCR-stimulated CTCLs, which are constitutively polarized in a Th2 phenotype in vivo2. We hypothesized that ZEB1 loss may contribute to GATA-3 overexpression in stimulated T cells. siRNA or shRNA knock down of ZEB1 in stimulated Jurkat cells caused significantly increased GATA-3 expression (Fig. 3e, Supplementary Figure 12).
Changes in expression of genes on recurrent SCNVs
We evaluated changes in gene expression of genes that reside in SCNVs. Among the 11 CTCL samples with RNA-Seq data, 3,752 genes were deleted a mean of 2.22 times (total of 8,317 gene deletions). We compared RNA transcript levels (fragments per Kb per million reads, FPKM) of each gene in haploid versus diploid states. We found that 87% of genes showed haploid transcript levels lower than the diploid mean (7224 of 8317; P < 10−300, chi-square) (Fig. 3f). There were 19 deletions in 7 tumor suppressor genes and 9 amplifications in 2 genes in this set and RNA-Seq data for at least 3 diploid tumors at each of these loci (Fig. 3g). Transcription of 18 of 19 deleted genes showed expression below the mean of diploid CTCLs (P= 9.6 × 10−5, chi-square), with mean FPKM of 55% of the non-deleted value. Similar results were observed for amplifications, with all 9 amplifications showing expression above the mean (P=0.003, chi-square, with mean expression 169% of diploid levels). Analysis of individual driver genes that had at least 3 diploid CTCLs and 3 CTCLs with SCNVs demonstrated statistically significant lower transcript levels in all 4 deleted genes (ARID1A, P=0.016; CTCF, P=0.042; TNFAIP3, P=0.012; ZEB1, P=0.024). Similarly, all 8 CTCLs with STAT5B amplifications had significantly increased transcript levels of STAT5B in (P=0.012) (Supplementary Table 15).
High frequency of pathogenic SCNVs
The data revealed a striking bias for SCNVs as drivers of CTCL. There were 12 statistically significant chromosome arm SCNVs and 36 significant focal SCNVs; these collectively occurred 473 times in CTCLs (mean 7.5 focal deletions, 1.6 broad deletions, 1.0 focal amplifications, 1.8 broad amplifications per CTCL; Figs. 1–3, Supplementary Figs. 4, Supplementary Tables 7–10). In contrast there were a total of only 38 SSNVs in CTCL drivers (1.0 per tumor; Supplementary Table 5). Thus, 92% of driver mutations in these CTCLs were SCNVs. This distribution sharply contrasts with tumor suppressors in other malignancies. For example, TP53 was deleted or mutated in 92.5% of CTCLs with a SCNV/SSNV ratio of 5.1:1, while in 17 other cancers with available data the mean ratio was 0.9:1 (range 0.08 – 2.0:1). Similarly, the SCNV/SSNV ratio for DNMT3A was 3.75:1 in CTCL versus 0.1:1 in AML (Fig. 4a). Further, ARID1A was deleted in 23 CTCLs, with only two SSNVs. Similar patterns were seen for ATM, CDKN2A, FAS, and ZEB1 (Figs. 3a). Collectively, for the 9 tumor suppressors identified (Fig. 1b, Table 2, Supplementary Table 14), somatic deletions accounted for 88% of the mutated alleles.
Figure 4. Contribution of SCNVs to CTCL.
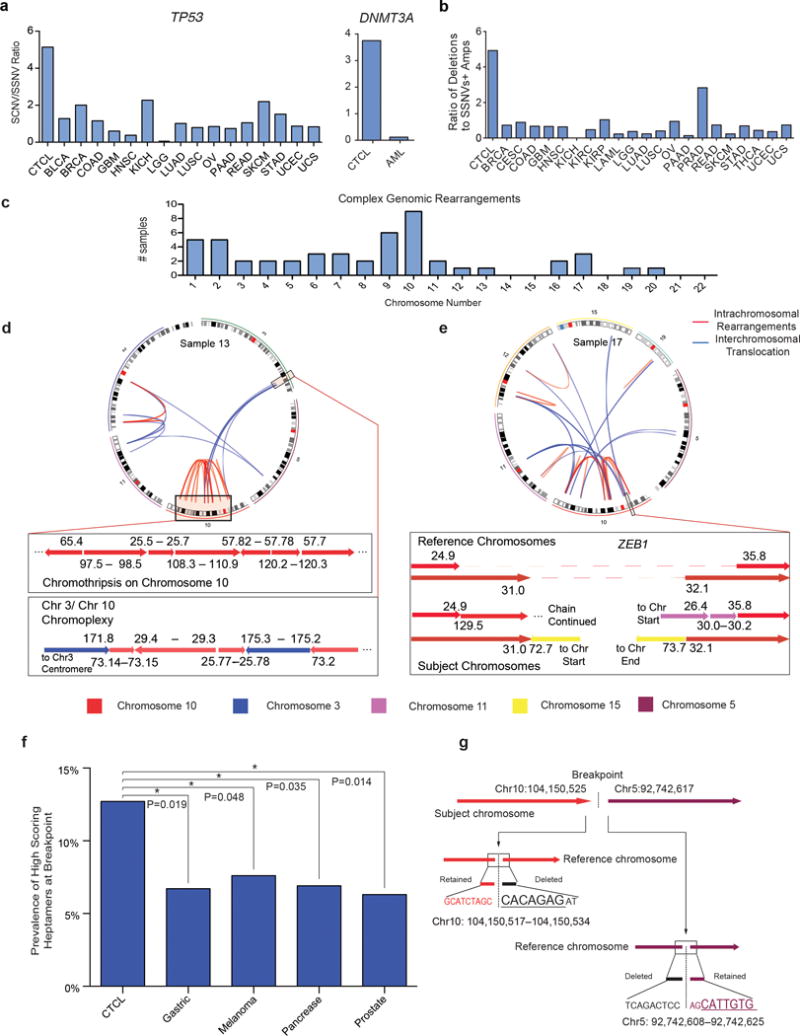
(a) Ratio of deletions: SSNVs in CTCL compared to other cancer-types for TP53 and DNMT3A. (b) Ratio of significant focal deletions to significant SSNVs + significant focal amplifications for driver mutations in CTCL compared to all cancers with available data from the TCGA. All cancers are annotated with the abbreviations assigned by the TCGA. (c) Number of CTCLs with complex genomic rearrangements at each chromosome. (d–e) CIRCOS plots and magnified views of structural rearrangements in two CTCLs. In the CIRCOS plots, blue lines represent interchromosomal translocations. Red lines represent intrachromosomal rearrangements. In the magnified views of individual structural variants, each arrow represents a chromosomal segment with the arrowhead indicating the direction of increasing nucleotide position number (hg18). The numbers bordering each arrow reflects the nucleotide positions of each contiguous genomic segment. The colors of the arrows reflect the chromosome from which it is derived. The reference chromosome is the wild-type chromosome. The dotted line in the reference chromosome represents regions of genomic loss in CTCL. Subject chromosomes reflect the rearranged chromosome in the CTCL sample. (f) High-scoring heptamers are enriched in the 55 bp windows at the breakpoints in CTCLs vs. epithelial cancers. High-scoring heptamers were defined as heptamers that are highly homologous to consensus RAG cleavage sites (RSS-score >8.55; Methods). * P-value<0.05, one-sided Fisher’s exact test. (g) High-scoring heptamers are present within 5 bp of the breakpoints underlying the C-terminal NFKB2 truncation in sample 17. The high-scoring heptamers are underlined.
We further compared the relative frequencies of pathogenic SCNVs and SSNVs in CTCL and 21 other cancers with publically available data from The Cancer Genome Atlas (TCGA) (see Methods). The ratio of significant focal deletions: (significant focal amplifications + SSNVs) in CTCL was 4.8:1, compared to a mean of 1:1 for 21 other cancers (range 0 – 2.7:1; Fig. 4b). Given that 18 genes are deleted 8–23 times while no gene has more than 7 SSNVs with 7 and no other gene statistically implicated in any cancer had more than 4 SSNVs, this ratio will not be altered by sequencing larger cohorts of CTCLs.
High frequency of complex structural rearrangements
The molecular events producing deletions were obscure from exome data. These were elucidated by whole genome sequencing of two CTCLs which identified 51 precise breakpoints contributing to 39 deletion events identified by exome sequencing (Supplementary Table 16, Supplementary Fig. 13,14). 39% of somatic deletions identified resulted from two or more chromosomal translocations with loss of intervening genomic DNA. 31% and 21% of deletions resulted from intrachromosomal deletions and chromosomal inversions, respectively.
Strikingly, 79% of these deletions were associated with complex structural rearrangements (Supplementary Table 16). Utilizing previously employed definitions of chromothripsis (evidence of more than 10 copy number states on a single chromosome45) we found that 65% of CTCL samples had evidence of at least one chromothripsis-like rearrangement (Supplementary Fig. 15, Fig. 4c). Interestingly, these events occurred most frequently on chromosomes harboring multiple CTCL tumor suppressors, allowing concurrent focal deletion of genes not closely linked on the same chromosome. For example, chromosome 10 showed complex rearrangement in 9 CTCLs (22.5%), producing deletion of ZEB1 (9 CTCLS), FAS (4 CTCLs), and C-terminal deletions of NFKB2 (3 CTCLs) (Fig. 4c–e). Similarly, ARID1A (4 CTCLs) and DNMT3A (5 CTCLs) are frequently deleted by chromothripsis-like events on chromosomes 1 and 2 (Fig. 4c). These results are distinct from the distribution of chomothripsis events identified by TCGA’s pan-cancer analysis, which reported the highest incidence on chromosomes 6, 9, and 12 with few chromosome 10 events46.
Enrichment of RAG heptamers in CTCL breakpoints
Because of the high incidence of narrow deletions (median size of focal deletions=1.6 Mb), we considered that breakpoints might often result from endonuclease-mediated cleavage by RAG genes (RAG1 and RAG2). RAGs initiate V(D)J recombination in developing lymphocytes by cleaving relatively well-conserved heptamer and nonamer elements. Recently, RAGs were implicated in recurrent deletions in ETV6-RUNX1-positive acute lymphoblastic leukemias47, with consensus RAG heptamers found in a 55 base pair window flanking deletion breakpoints in 40% of cases47. Using the same algorithm (see Methods), we found consensus RAG heptamers flanking 12.7% of CTCL deletion breakpoints (Supplementary Figs. 16a,b). This was significantly higher than seen for deletion breakpoints in epithelial cancers with available data (P<0.05, Fisher’s test) (Fig. 4f, Supplementary Fig. 16b).
We examined recurrent structural variants for high scoring RAG cleavage sites since RAGs have been implicated in several recurrent translocations48. From our WGS, we found a high scoring heptamer at both ends of the intragenic NFKB2 translocation in CTCL 17 (Fig. 4g). We identified from the literature four additional B-cell lymphomas with defined breakpoints leading to C-terminal truncations of NFKB29. These five breakpoints all clustered within a 1 kb segment. Two harbored high-scoring heptamers (RSS>8.55) within the 55 base pairs flanking the breakpoint (P=0.005; binomial distribution) (Supplementary Fig. 16c). A third breakpoint harbored a heptamer with an RSS of 7.55; 13.6% of rearrangements at the endogenous human TCR locus have RSS scores between 7.5 and 8.55 (Supplementary Fig. 17). The presence of heptamers with an RSS > 7.5 in three of five NFKB2 breakpoints is unlikely to occur by chance (P=0.003; binomial distribution) (Supplementary Fig. 16c).
Discussion
These findings implicate somatic mutations in 17 genes in CTCL. We find frequent deletions and damaging SSNVs in chromatin-modifying genes [ARID1A (62.5%), CTCF (12.5%), and DNMT3A (42.5%)]. Many genes mutated in CTCL also contribute to other T cell neoplasms, including peripheral T cell lymphoma (CD2824, DNMT3A49, RHOA22–24), T-large granular lymphocytic leukemia (STAT5B26), and adult T cell leukemia/lymphoma (ZEB142), underscoring the importance of these genes for malignant transformation of mature T cells. Consistent with this, we found mutations in multiple components of the TCR signaling pathway, including CD28, TCR-associated enzymes (PLCG1, PRKCQ, TNFAIP3), and transcription factors (NFKB2, STAT5B, ZEB1). We found mutations in genes that drive Th2 differentiation (ZEB1), that facilitate escape from TGF-β-mediated growth suppression42 (ZEB1), and that facilitate resistance to TNFRSF-mediated apoptosis50 (FAS and ARID1A).
We identified an additional 23 significant focal SCNVs whose length precluded the confident identification of single gene drivers. Eight of these contain one or more consensus cancer genes (e.g. RB151, deleted in 25% of CTCLs) (Supplementary Table 12). In eight additional intervals, GRAIL analysis suggested candidate target genes (Supplementary Tables 12,13), including PDCD1 (biallelically deleted in 5% of CTCLs, heterozygously deleted in 15%; GRAIL P-value= 0.0035), CARD11 (amplified in 22.5% of CTCLs, GRAIL P-value= 0.0003) and JAK2 (amplified in 12.5% of CTCLs, GRAIL P-value=3.27 × 10−5). PDCD1 prevents continuous T cell proliferation in the setting of chronic TCR engagement52. CARD1153 and JAK254,55 are known oncogenes in hematological malignancies, encoding critical components of the NF-κB56 and STAT554 signaling pathways, respectively. Among the two remaining narrow intervals (3 genes or fewer), GRAIL identified LATS1 (deleted in 22.5%; P-value=0.17) and ZNF365 (deleted in 50%; GRAIL P-value=0.58) as the most likely target genes (Supplementary Table 12). Lastly, we identified twelve significant broad SCNVs including chromosome 8q amplifications (42.5% of CTCLs; Q =3.9×10−11) which occur in many cancers19 and contain the MYC oncogene57 (Supplementary Table 10).
The genomic architecture of CTCL is unusual in having a large contribution of focal deletions (7.5 significant focal deletions/sample) (Figs. 3a,b, Fig. 4a,b). The possible involvement of RAGs in these events is surprising given that CTCLs are thought to derive from RAG-negative mature T cells. Additional WGS will be necessary to further evaluate the link between RAGs and genome instability in CTCL.
Our findings establish CD28 as an oncogene. Recurrent p.Phe51Val and p.Gln77Pro mutations increase CD28 avidity for CD86, which enhances downstream signaling. A single instance of the p.Phe51Val mutation was recently reported in angioimmunoblastic T cell lymphoma, lending further support to this recurrent mutation24. The increased avidity conferred by these SSNVs are similar to those resulting from amino acid substitutions in the second generation CTLA-4-Fc fusion protein, belatacept58. A recent report in peripheral T cell lymphomas identified another recurrent SSNV in CD28 in the intracellular domain (p.Thr195Pro)24. We posit this mutation may also augment CD28 signaling.
These findings have potential therapeutic implications. Frequent mutations activating the NF-κB pathway (NFKB2 truncations and TNFAIP3) suggest potential utility of NF-κB inhibitors such as bortezomib; a recent small phase II clinical trial supports this possibility59, and suggests evaluation of the correlation of mutation in these genes with response to therapy. Similarly, CD28 mutations suggest therapeutic potential for inhibitors of binding such as abatacept (CTLA-4-Ig), which is used in autoimmune disease60. Lastly, CTCL responds to histone deacetylase inhibitors61, raising the question of whether the frequent chromatin modifying gene mutations account for this sensitivity. The genomic landscape of CTCL defined herein defines the therapeutic opportunities and challenges for the future.
Online methods
Sample selection and preparation
This study was reviewed and approved by the human subjects institutional review boards (IRBs) of the Yale School of Medicine and the Johns Hopkins University School of Medicine. Written informed consent was obtained from all living participants. We chose to perform whole exome sequencing on leukemic CTCL from 40 patients. The patients all had Stage IVA-B CTCL with a detectable abnormal population in the blood. The leukemic cells were isolated using flow cytometry activated cell sorting using cell surface markers that uniquely identified the neoplastic clones. If the TCRVβ antibody was available, we isolated the CD3+ TCRVβ+ CD14− CD8− CD19− population. If not, we isolated CD3+ CD26− CD14− CD8− CD19− cells. We found the mutation spectrum of cells were similar regardless of method of isolation. For the normal controls, we isolated the CD14+ CD3− CD8− CD19− monocytes. The median purity of the CTCL tumor cells and matched normal monocytes was 98% (Supplementary Fig. 1). None of the patients had concomitant acute myelogenous leukemia or myelodysplasia. All antibodies were from BD Biosciences. DNA was extracted from the cells using standard techniques (Qiagen).
Sequencing
For whole exome sequencing, DNA libraries were prepared by Covaris sonication, size selected, and ligated to specific barcoded adaptors (Illumina TruSeq) for multiplexed analysis. Targeted capture was performed using the NimbleGen 2.1 Exome reagent, followed by sequencing on Illumina HiSeq system. All samples achieved >140× coverage in exon regions. Sequences were aligned to the human genome build 36 with ELAND (Illumina).
Somatic mutation calling and identification of significantly mutated genes
The significance of differences in read distributions between tumor and monocyte sequencing data were evaluated at all covered positions, using a two-tailed Fisher’s exact test. P-value ranked lists of somatic calls identified in the tumor were compared to variant calls unique to blood by the same analysis and a P-value significance threshold was determined independently for each tumor to yield a list of high-confidence somatic calls. Somatic mutations were filtered to remove variants present in public and Yale databases, which are likely miscalled germline variants. Previous studies have demonstrated the inverse relationship between gene expression and mutation rate13. To define the RNA levels of genes in CTCL, we utilized the previously described data set5. We confirmed in our data set an inverse relationship between gene expression and mutation rate (R2 = −0.90604). Significantly mutated genes were identified using the previously described approach, the convolution test (CT)62. CT calculates a summarized log statistic of joint binomial point probability, as first proposed by Getz et al63. Significant recurrent missense mutations were identified as previously described64. For known recurrent somatic mutations implicated in CTCL or other cancers, the probability of recurrence in the CTCL set by chance was calculated from the length of the coding region, total number of known recurrent sites in the gene and the total number of observed somatic mutations in the CTCL set.
Somatic copy number mutations
Somatic copy number data was generated from WES data from the 40 matched samples by utilizing coverage depth ratio and changes in B-allele frequency (BAF) of informative SNPs between each tumor and matched normal sample. To identify SCNVs, the coverage ratio was first normalized by GC content and overall coverage depth of the exome using CONTRA65. Contiguous chromosomal segments with similar copy number were merged. The SCNVs were confirmed by ΔBAF. Segments with discordant ΔBAF and coverage depth were discarded for the analysis. We defined focal SCNVs as chromosomal segments <0.5 chromosome arm, broad SCNVs >0.5 chromosome arm. Homozygous deletions were defined as SCNVs with a mean LRR<−2.0. For the sake of clarity, SCNVs at the TCR gene loci, which are deleted in all mature CD4+ T cells, were manually excluded from the analysis. To identify the events which occur more often than expected by chance, we utilized GISTIC 2.014. To identify genes in recurrent focal lesions, we utilized the following settings: arbitrated peel-off, confidence interval=90%, false discovery rate <0.25. To identify candidate target genes, we first identified genes with significant gene-localizing mutations (see below). We then identified consensus cancer genes identified to be significantly mutated by the cancer genome atlas (TCGA) across multiple tissue types17. When possible, we utilized tumor suppressors as the target gene on recurrent deletions, and oncogenes as the target gene on recurrent amplifications. For the remaining intervals, we performed GRAIL analysis (see below) to identify the target gene. The lone exception is TNFAIP3, which was recently implicated in leukemic CTCL10.
Identification of specific genes in significant SCNV intervals
For each significant SCNV (focal and broad), the probability of the observed number of focal SCNVs and protein-altering SSNVs occurring by chance in each gene was determined from the binomial distribution, and Q values were calculated using Bonferroni correction for 20,025 protein-coding genes to produce a family-wise error rate. The likelihood ratio favoring the most likely gene over then next most likely gene in each interval was calculated from the reciprocal of the ratio of their P-values. Focal deletions were attributed to a gene if the deletion included at least one coding exon of the gene. Amplifications were analyzed similarly except the entire coding region had to be encompassed by the SCNV to be included. Genes with a likelihood ratio >1000:1 were considered significant.
GRAIL analysis
We utilized the GRAIL algorithm to identify target genes in focal SCNVs that are functionally related to driver genes identified by gene-localizing mutations. The full methods and algorithm for GRAIL18 is available at, using the default settings. All PubMed abstracts were queried until October 2014. The genes identified by gene-localizing mutations were ARID1A, BRAF, CDKN2A, CD28, DNMT3A, FAS, NFKB2, PLCG1, RHOA, STAT5B, TP53, and ZEB1. The significant focal amplifications and deletions were queried together.
Whole genome sequencing
CTCL DNA for 2 cases was prepared for short-insert (500-bp) library construction flow-cell preparation and cluster formation using the Illumina no-PCR protocol66. We performed 150-base paired-end sequencing on an Illumina HiSeq 2000. Sequenced reads were aligned to the human genome (NCBI Build 37) using the Burrows-Wheeler aligner (BWA) with default settings67.
Somatic copy number mutations from WGS
SCNVs were identified as segments with altered coverage depth and allelic imbalance using Pathwork-R68. As described previously, the genome is segmented into 50 kb bins, coverage is normalized for GC content, and segments of similar copy number are joined by circular binary segmentation. Simultaneously, single-nucleotide variants are extracted using SAMtools and discovered variants are filtered using a list of known SNPs (dbSNP). Segments are assigned a copy number based on coverage depth and allelic imbalance (Supplementary Fig. 2).
Breakpoint Identification
We identified anomalous paired end reads supporting a structural variant utilizing Breakdancer as previously described69. To resolve the breakpoint at the base-pair resolution, we queried the fastq files for the soft- and hard-clipped reads at each breakpoint. Complex genomic rearrangements were defined as requiring more than the minimal number of breakpoints for the structural variant, i.e. 2 breakpoints for a deletion or inversion and 4 for a translocation event. These include unbalanced translocations, i.e. requiring the involvement of 3+ chromosomes, such as chromoplexy (multiple contiguous translocations between two or more chromosomes) or chromothripsis (deletions or inversions without loss of all of the genomic regions between the breakpoints; intervening DNA is inserted into another genomic region).
RNA-Sequencing
mRNA from 11 samples were isolated, extracted, and subject to sequencing on Illumina HiSeq 2000 as previously described70. There were a median of 87 million unique reads per sample. The RNA was aligned and quantified by fragments per kilobase per million reads using the Tuxedo suite as previously described71. Transcript levels (fragments per kilobase per million reads) were assessed in CTCLs with and without SCNVs. P-values were calculated using the chi-square test of the null hypothesis that a gene in an SCNV has an equal probability of being expressed higher or lower than the mean of the diploid value. One-sided P-values are reported for deletions showing lower expression and amplifications showing higher expression. For analysis of expression of individual genes, expression of genes with RNAseq data from at least 3 diploid and 3 SCNV tumors were analyzed by Mann-Whitney test.
TCGA Analysis
We downloaded the data matrices from The Cancer Genome Atlas on 02-15-2014. On 02-05-2015, the data for each cancer in the initial analysis was updated. For analysis of individual genes, we counted the number of SSNVs and number of deletions (focal and broad) for each gene. For the global analysis, we did the following. We summed the SSNVs that occur on genes determined by MutSig to have a false discovery rate (FDR) or Q-value <0.25. Loci for significant focal deletions and focal amplifications were determined by GISTIC with a false discovery rate (FDR) or Q-value<0.25. We then summed the number of times each significant deletion GISTIC confidence interval was focally deleted. Similar analysis was performed for the amplifications. These sums were used to calculate the ratio of significant focal deletions vs. the sum of SSNVs and significant focal amplifications.
Identification of RAG heptamers
As described previously, we utilized FIMO to identify high scoring heptamers72. We used a similar window at the breakpoint assuming a resection of −5 to 50 bp from the heptamer to the breakpoint47. We evaluated incidence of high scoring heptamers in CTCL vs. other cancers utilizing the one-sided Fisher’s test. For the NFKB2 analysis, we generated a sliding window of 55 bp within 2 kb of the NFKB2 breakpoints. We calculated the prevalence of 55 bp windows harboring a heptamer with an RSS score of 8.55 or higher (0.08) and the prevalence of 55 bp windows harboring a heptamer with an RSS score of 7.5 or higher (0.16). The statistical enrichment of high-scoring heptamers in 55 bp windows at NFKB2 breakpoints was calculated by the binomial distribution. For the human T cell receptor analysis, we downloaded the reference RSS sequences from the human TCR gene loci from RSSsite73. RSS scores were calculated for these sequences using FIMO from the MEME suite72.
Structural modeling
Structural modeling of CD28 and its interactions with CD80 and CD86 was performed using PDB ID 1YJD (CD28)27 and PDB ID 1I85 (CTLA-4-CD86)20. CD28 coordinates were extracted from PDB ID 1YJD, and superimposed using secondary structure alignment with CTLA-4 using COOT software74 and energy minimized within UCSF Chimera (Resource for Biocomputing, Visualization, and Informatics, University of California, San Francisco). Figures were prepared using UCSF Chimera.
CD28 codon-based mutagenesis
The CD28 construct was obtained from DNASU. The p.F51V and p.Q77P mutations were introduced by Quickchange PCR (Stratagene). The wild-type CD28 and CD28 mutants were cloned into p-Lenti-TOPO according to manufacturer’s instructions (Life Technologies).
Flow cytometric analysis
293T (CRL-3216) and Jurkat T cells (E6.1) were obtained from ATCC (American Type Culture Collection). They were routinely checked for mycoplasma and were mycoplasma free. transduced with pLenti-TOPO containing wild-type CD28 and the CD28 mutants. CD28 expression was detected with a mouse anti-human CD28 (Clone 28.2; dilution 1:100; eBioscience). The binding of human CD80-Fc (catalog number 140-B1-100; R&D Systems) and human CD86-Fc (catalog number 141-B2-100; R&D Systems) to these cells were performed as previously described29,58. The secondary antibody used was Alexa Fluor 647 conjugated goat anti-human IgG secondary antibody (catalog number A-21445; concentration: 5 μg/ml; Life Technologies). Kd were calculated based on the presumption of one binding site. Kd= [A][B]/[AB] where in A represent soluble CD80-Fc or Cd86-Fc and B represents CD28 molecules on the cell surface of 293T cells.
Assay for IL-2 production in Jurkat T cells
Jurkat T cells were transduced with pLenti-TOPO containing containing wild-type CD28 and CD28 (p.Phe51Val). 300,000 Jurkat T cells were stimulated with phorbol myristate acetate (PMA) (Sigma) (50 ng/ml) and ionomycin (1 μg/ml) (Sigma) for 6 hours in the presence of indicated levels of CD86-Fc (R&D). RNA was prepared according to manufacturer protocols (Qiagen). IL-2 levels were assessed with quantitative PCR utilizing SybrGreen (BioRad) with the primers listed in Supplementary Table 17. GAPDH was used as a control with previously described primers75.
Generation of ZEB1-deficient Jurkat T cells
Jurkat cells were obtained from ATCC. They were maintained in RPMI 1640 media with L-glutamine supplemented with 10% FBS and antibiotics. 2 million Jurkat cells were transfected with 30pM Accell ZEB1 siRNA (A-006564-14) or scrambled control (Dharmacon) using the cell-line specific nucleofector settings (Lonza). 64 hours post nucleofection, cells were stimulated with PMA (20 ng/mL) and calcium Ionomycin (500 ng/mL). Alternatively, they were transduced with shRNA targeting ZEB1 (TRCN0000369266) or scrambled control.
Western blot analysis
Whole cell lysates were analyzed by western blot using ZEB1 (Clone D80D3; Cell Signaling; dilution: 1:1000), GATA3 (Clone D13C9; Cell Signaling; dilution 1:1000) and RAF1 (Clone 53/c-RAF-1; BD Transduction Laboratories; dilution 1:1000) antibodies with rabbit and mouse secondary antibodies (Santa Cruz; dilution 1:2500) respectively.
Supplementary Material
Acknowledgments
J.C. is supported by the Dermatology Foundation and the Yale SPORE in Skin Cancer Career Development Award. T.J.B. is supported by the Yale SPORE in Skin Cancer (P50 CA121974). G.G. is supported by the Agency for Science, Technology and Research, Singapore. M.G. is supported by the NIH grant RO1 CA102703. R.P.L. and D.G.S. are Investigators of the Howard Hughes Medical Institute.
Footnotes
URLs
TCGA, https://tcgadata.nci.nih.gov/tcga/dataAccessMatrix.html
GRAIL, www.broad.mit.edu/mpg/grail
Accession codes
The de-identified genetic whole genome sequencing, whole exome sequencing, and RNA sequencing data have been deposited to the Sequence Read Archive according to the guidelines outlined by the Yale Institutional Review Board. The accession code to this data is SRP058948.
Author contributions
J.C., M.G., and R.P.L. designed the experiments and wrote the manuscript. J.C. performed the majority of the experiments. J.C., G.G., and R.P.L. performed and analyzed sequencing. J.C., T.W., B.S.H., T.W., K.C., K.J.L., J.M.L., L.B., F.M.F., A.S., E.C.V., R.L.E., and M.G. collected and annotated clinical samples. C.G.B. and T.J.B. performed the structural modeling. B.S.H., K.C., T.W. helped with functional experiments. J.D.O. helped with the sample preparation. K.C., R.D.B., and J.T. helped with analysis. Y.M. and D.G.S. helped with the cryptic RAG sequence analysis. There are no competing financial interests.
References
- 1.Girardi M, Heald PW, Wilson LD. The pathogenesis of mycosis fungoides. N Engl J Med. 2004;350:1978–88. doi: 10.1056/NEJMra032810. [DOI] [PubMed] [Google Scholar]
- 2.Guenova E, et al. TH2 cytokines from malignant cells suppress TH1 responses and enforce a global TH2 bias in leukemic cutaneous T-cell lymphoma. Clin Cancer Res. 2013;19:3755–63. doi: 10.1158/1078-0432.CCR-12-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim EJ, et al. Immunopathogenesis and therapy of cutaneous T cell lymphoma. J Clin Invest. 2005;115:798–812. doi: 10.1172/JCI24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong HK, Mishra A, Hake T, Porcu P. Evolving insights in the pathogenesis and therapy of cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome) Br J Haematol. 2011;155:150–66. doi: 10.1111/j.1365-2141.2011.08852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin WM, et al. Characterization of the DNA Copy-Number Genome in the Blood of Cutaneous T-Cell Lymphoma Patients. J Invest Dermatol. 2011 doi: 10.1038/jid.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermeer MH, et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer Res. 2008;68:2689–98. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- 7.Caprini E, et al. Identification of key regions and genes important in the pathogenesis of sezary syndrome by combining genomic and expression microarrays. Cancer Res. 2009;69:8438–46. doi: 10.1158/0008-5472.CAN-09-2367. [DOI] [PubMed] [Google Scholar]
- 8.Laharanne E, et al. Genome-wide analysis of cutaneous T-cell lymphomas identifies three clinically relevant classes. J Invest Dermatol. 2010;130:1707–18. doi: 10.1038/jid.2010.8. [DOI] [PubMed] [Google Scholar]
- 9.Neri A, Fracchiolla NS, Migliazza A, Trecca D, Lombardi L. The involvement of the candidate proto-oncogene NFKB2/lyt-10 in lymphoid malignancies. Leuk Lymphoma. 1996;23:43–8. doi: 10.3109/10428199609054800. [DOI] [PubMed] [Google Scholar]
- 10.Braun FC, et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sezary syndrome. Leukemia. 2011;25:1494–501. doi: 10.1038/leu.2011.101. [DOI] [PubMed] [Google Scholar]
- 11.Vaque JP, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood. 2014 doi: 10.1182/blood-2013-05-504308. [DOI] [PubMed] [Google Scholar]
- 12.Wong HK. Novel biomarkers, dysregulated epigenetics, and therapy in cutaneous T-cell lymphoma. Discov Med. 2013;16:71–8. [PubMed] [Google Scholar]
- 13.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedwick CE, Altman A. Perspectives on PKCtheta in T cell activation. Mol Immunol. 2004;41:675–86. doi: 10.1016/j.molimm.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Man K, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–65. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 22.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–70. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–5. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 24.Yoo HY, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:371–5. doi: 10.1038/ng.2916. [DOI] [PubMed] [Google Scholar]
- 25.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajala HL, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121:4541–50. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans EJ, et al. Crystal structure of a soluble CD28-Fab complex. Nat Immunol. 2005;6:271–9. doi: 10.1038/ni1170. [DOI] [PubMed] [Google Scholar]
- 28.Odorizzi PM, Wherry EJ. Inhibitory receptors on lymphocytes: insights from infections. J Immunol. 2012;188:2957–65. doi: 10.4049/jimmunol.1100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linsley PS, et al. Human B7-1 (CD80) and B7-2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity. 1994;1:793–801. doi: 10.1016/s1074-7613(94)80021-9. [DOI] [PubMed] [Google Scholar]
- 30.Slavik JM, Hutchcroft JE, Bierer BE. CD80 and CD86 are not equivalent in their ability to induce the tyrosine phosphorylation of CD28. J Biol Chem. 1999;274:3116–24. doi: 10.1074/jbc.274.5.3116. [DOI] [PubMed] [Google Scholar]
- 31.Wang K, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 32.Kakiuchi M, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–7. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 33.Mayer T, Meyer M, Janning A, Schiedel AC, Barnekow A. A mutant form of the rho protein can restore stress fibers and adhesion plaques in v-src transformed fibroblasts. Oncogene. 1999;18:2117–28. doi: 10.1038/sj.onc.1202537. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Mata R, et al. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 2006;406:425–37. doi: 10.1016/S0076-6879(06)06031-9. [DOI] [PubMed] [Google Scholar]
- 35.Legarda-Addison D, Ting AT. Negative regulation of TCR signaling by NF-kappaB2/p100. J Immunol. 2007;178:7767–78. doi: 10.4049/jimmunol.178.12.7767. [DOI] [PubMed] [Google Scholar]
- 36.Kopp KL, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013;12:1939–47. doi: 10.4161/cc.24987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, et al. Cutaneous T cell lymphoma expresses immunosuppressive CD80 (B7-1) cell surface protein in a STAT5-dependent manner. J Immunol. 2014;192:2913–9. doi: 10.4049/jimmunol.1302951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haddad BR, et al. STAT5A/B gene locus undergoes amplification during human prostate cancer progression. Am J Pathol. 2013;182:2264–75. doi: 10.1016/j.ajpath.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly J, et al. A role for Stat5 in CD8+ T cell homeostasis. J Immunol. 2003;170:210–7. doi: 10.4049/jimmunol.170.1.210. [DOI] [PubMed] [Google Scholar]
- 40.Wu JN, Roberts CW. ARID1A Mutations in Cancer: Another Epigenetic Tumor Suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132:745–54. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 42.Hidaka T, et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood. 2008;112:383–93. doi: 10.1182/blood-2008-01-131185. [DOI] [PubMed] [Google Scholar]
- 43.Williams TM, et al. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–4. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 44.Gregoire JM, Romeo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. J Biol Chem. 1999;274:6567–78. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 45.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014 doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gostissa M, et al. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature. 2009;462:803–7. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–6. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 50.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Fiore R, D’Anneo A, Tesoriere G, Vento R. RB1 in cancer: different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. J Cell Physiol. 2013;228:1676–87. doi: 10.1002/jcp.24329. [DOI] [PubMed] [Google Scholar]
- 52.Yao S, Zhu Y, Chen L. Advances in targeting cell surface signalling molecules for immune modulation. Nat Rev Drug Discov. 2013;12:130–46. doi: 10.1038/nrd3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer SC, Levine RL. Molecular pathways: molecular basis for sensitivity and resistance to JAK kinase inhibitors. Clin Cancer Res. 2014;20:2051–9. doi: 10.1158/1078-0432.CCR-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32:2601–13. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 56.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002;21:5184–94. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsen CP, et al. Rational development of LEA29Y (belatacept), a high-affinity variant of CTLA4-Ig with potent immunosuppressive properties. Am J Transplant. 2005;5:443–53. doi: 10.1111/j.1600-6143.2005.00749.x. [DOI] [PubMed] [Google Scholar]
- 59.Zinzani PL, et al. Phase II trial of proteasome inhibitor bortezomib in patients with relapsed or refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:4293–7. doi: 10.1200/JCO.2007.11.4207. [DOI] [PubMed] [Google Scholar]
- 60.Genovese MC, et al. Abatacept for rheumatoid arthritis refractory to tumor necrosis factor alpha inhibition. N Engl J Med. 2005;353:1114–23. doi: 10.1056/NEJMoa050524. [DOI] [PubMed] [Google Scholar]
- 61.Rangwala S, Zhang C, Duvic M. HDAC inhibitors for the treatment of cutaneous T-cell lymphomas. Future Med Chem. 2012;4:471–86. doi: 10.4155/fmc.12.6. [DOI] [PubMed] [Google Scholar]
- 62.Dees ND, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–98. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Getz G, et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 64.Zhao S, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci U S A. 2013;110:2916–21. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–13. doi: 10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozarewa I, Turner DJ. Amplification-free library preparation for paired-end Illumina sequencing. Methods Mol Biol. 2011;733:257–66. doi: 10.1007/978-1-61779-089-8_18. [DOI] [PubMed] [Google Scholar]
- 67.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayrhofer M, Dilorenzo S, Isaksson A. Patchwork: allele-specific copy number analysis of whole-genome sequenced tumor tissue. Genome Biol. 2013;14:R24. doi: 10.1186/gb-2013-14-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goh G, et al. Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors. Nat Genet. 2014;46:613–7. doi: 10.1038/ng.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merelli I, et al. RSSsite: a reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res. 2010;38:W262–7. doi: 10.1093/nar/gkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 75.Choi J, et al. Identification of PLX4032-resistance mechanisms and implications for novel RAF inhibitors. Pigment Cell Melanoma Res. 2014;27:253–62. doi: 10.1111/pcmr.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


