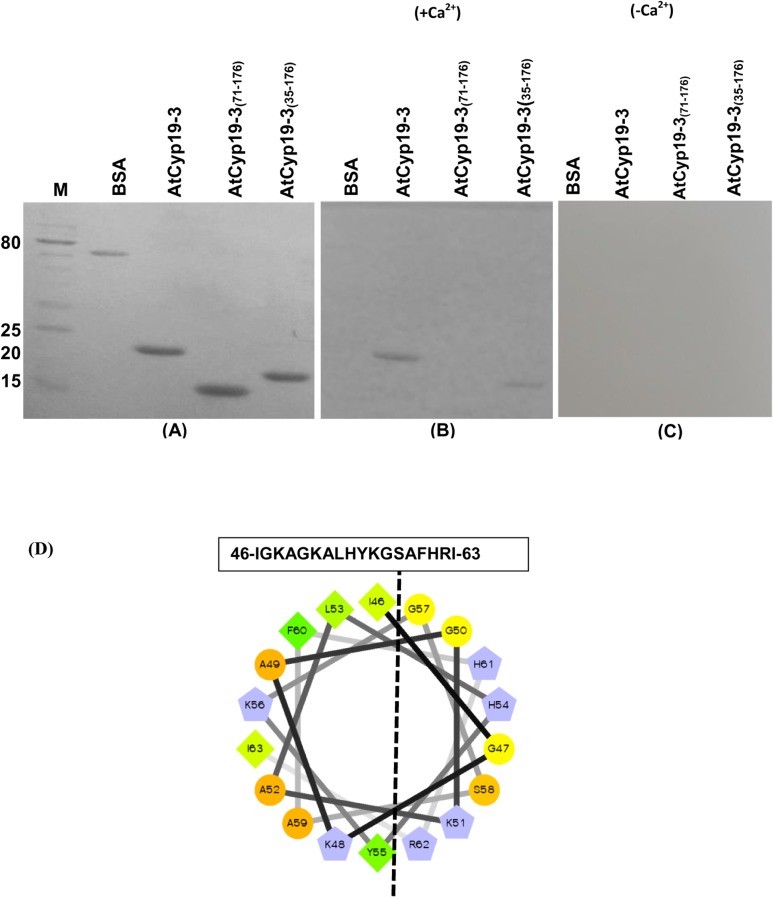
Fig 7. Calmodulin (CaM) gel-overlay assay of the purified recombinant AtCyp19-3 and its truncated versions.
The purified proteins were resolved by using 15% SDS-PAGE followed by transfer on to Hybond C nitrocellulose membrane. Proteins transferred on to membrane, after Ponceau S staining (A), were allowed to renature in the presence (B) and absence of Ca2+ (C) for 16 h at 4°C. The membrane was incubated with biotinylated-CaM in the presence (+Ca2+) and absence of Ca2+(-Ca2+), followed by probing with streptavidin-alkaline phosphate conjugate. CaM-binding proteins were visualized using nitro-blue tetrazolium/5-bromo-4-chloro-3-indolyphosphate as substrate. (D) Helical wheel projection (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi) of the predicted CaM-binding domain (CaMBD) of AtCyp19-3. The dashed line separate the hydrophilic and the hydrophobic faces of the amphiphilic CaMBD. Hydrophilic residues: circles; hydrophobic residues: diamonds; potentially negatively charged: triangles; potentially positively charged: pentagons. Hydrophobic residues are depicted in green and the amount of green decreases proportionally to the hydrophobicity, with zero hydrophobicity coded as yellow. Hydrophilic residues are shown as red, with the amount of red being proportional to the hydrophilicity. Light blue represents the potentially charged residues. M-protein markers (kDa).