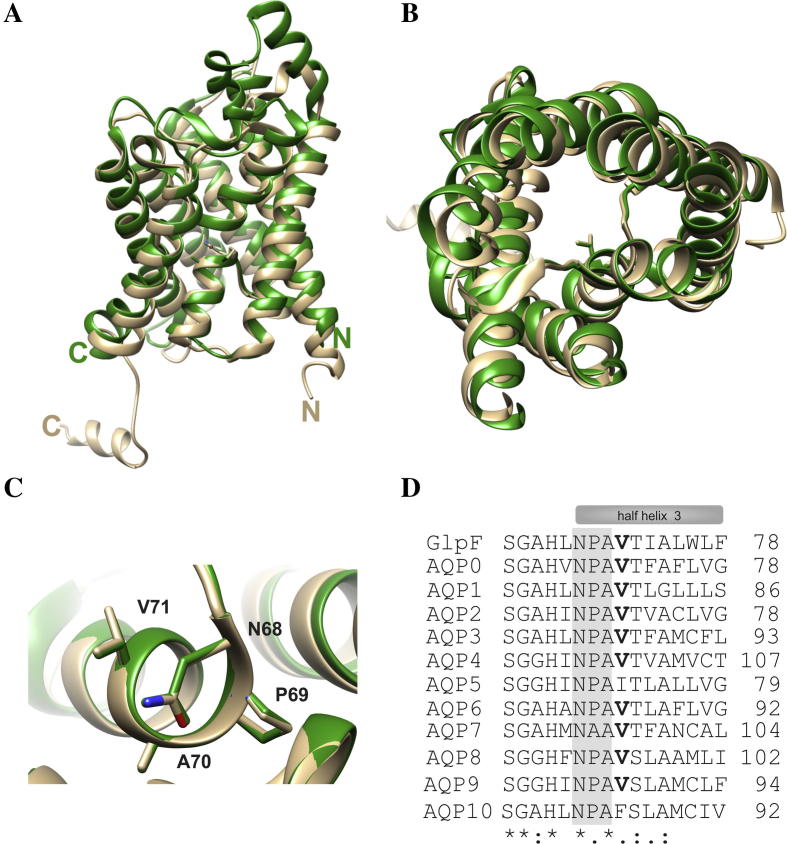
Fig. 1.
Alignment of the GlpF and AQP2 structures and sequences. Superposing the structures of GlpF (green) and AQP2 (beige) illustrates the high conservation of the AQP tertiary structures. While structural differences are found in the extra membranous loops in the crystal structures, the conformation of the TM helices is conserved. (A) Superposition of GlpF and AQP2 seen sideways. Both, the N- and the C-terminus, are located in the cytoplasm. (B) Top view onto the translocation pore as seen from the periplasm or extracellular space, respectively. Depicted is the amino acid residue Val71 in GlpF and in AQP2. For clarity, the periplasmatic/extracellular loop regions are not shown. (C) Zoom in on the structure around the first NPA motif. The structural alignment was prepared using the program Chimera [29]. (PDB: 1FX8 und 4NEF). (D) Sequence alignment of GlpF and the classical human AQPs and aquaglyceroporins. The conserved NPA motif is highlighted in gray, and the conserved Val residue (position 71 in AQP2) is highlighted in bold. One dot highlights conserved residues, two dots highly conserved residues and a star indicates identical residues. The sequence alignment was performed using the program ClustalW2 [30]. The sequences were obtained from the UniProtKB/TrEMBL data bank [31].