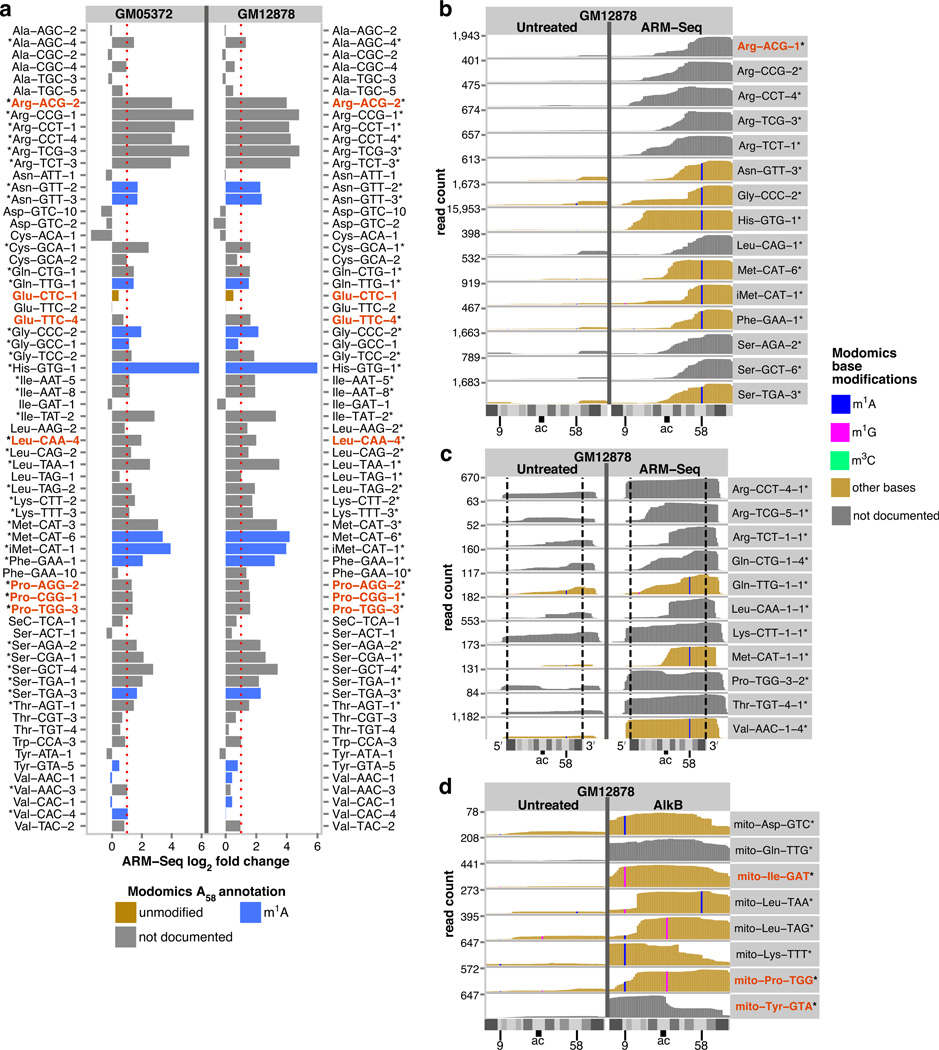
Figure 3. ARM-Seq reveals methylated RNAs derived from human cytosolic tRNAs, tRNA precursors, and mitochondrial tRNAs.
(a) Transcriptome-scale screening using ARM-Seq provides evidence for m1A58 modification in a majority of human tRNA isotypes, showing a consistent profile of modification in two B-cell derived human cell lines (with * indicating significant responders). (b) Profiles for many tRNA-derived small RNAs revealed by ARM-Seq show little, if any detection in untreated samples, indicating high levels of modification. (c) ARM-Seq also provides the first evidence that many human pre-tRNAs are m1A58 modified at an early stage prior to removal of 5’ leader and 3’ trailer sequences from primary transcripts (demarcated by dashed lines), demonstrating the ability to resolve sequential modification and processing steps involved in tRNA maturation. The 5′-leader sequences of these precursor-derived RNAs were typically short (4–5 nt) when present, which might reflect either nucleolytic processing or dephosphorylation of triphosphorylated primary transcripts to generate 5’-monophosphate ends (required for RNA-seq library inclusion). By contrast, the 3′-trailers were often 9–10 nt or longer, frequently ending with a poly-U sequence, suggesting that these represent the 3′-ends of primary RNA polymerase III transcripts. Reads for full-length and fragmentary pre-tRNAs revealed by ARM-Seq included the T-loop region, consistent with m1A58 modifications. (d) Fragments of human mitochondrial tRNAs revealed by ARM-Seq demonstrate a capacity to also demethylate m1A9 (in mito-Asp-GTC, mito-Lys-TTT), m1G9 (mito-Ile-GAT), and m1G37 (mito-Leu-TAG, mito-Pro-TGG), enabling investigation of mitochondrial diseases related to tRNA modification and processing. tRNAs for which ARM-Seq predictions were verified by primer extension are indicated in red.