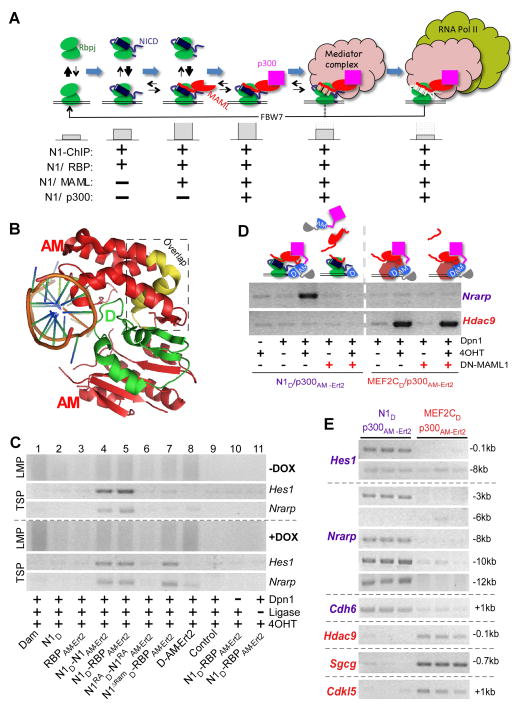
Figure 1. SpDamID Proof-of-Concept Utilizing the Notch Signaling Pathway (see also Figure S1).
(A) The step-wise assembly of Notch transcriptional complexes. The hypothetical stability of the complex on DNA is indicated by the size of the gray bars below. Notch1 (N1) ChIP probes regions bound by all the complexes. SpDamID pairs identify only regions bound by a specific complexes (marked with +). (B) Crystal structure of Dam. The N-terminal D half (green), C-terminal AM half (red), and overlapping region (yellow) are shown. (C) DNA from mK4 cells expressing various proteins amplified by Ligation mediated PCR (panels labeled LMP, showing a library aliquot) or target specific PCR (panels labeled TSP, showing semi-quantitative PCR for Hes1 or Nrarp enhancer). Panels above the dotted line record enrichment from proteins expressed without DOX; panels below the dotted line record enrichment with DOX induced expression of the following proteins: Lane1, full-length Dam; Lane 2–3, individual halves of SpDam fused to either Notch (lane 2) or RBP (lanes 3); Lanes 4–7, co-expression of SpDam pairs fused to various Notch complex components or mutants as indicated. Lane 8, co-expression of unfused SpDam halves. Lane 9, non-transfected control. Lane 10, no DpnI and Lane 11, no ligase. All experiments were performed in the presence of 4OHT. (D) Expression of DN-MAML1 prevents recruitment of p300 to Notch, and thus blocks labeling of Nrarp by N1D/p300AM-Ert2. DN-MAML1 does not interfere with enrichment of Hdac9 by Mef2cD/p300AM-Ert2. (E) Target-specific PCRs on biological triplicate samples labeled by N1D/p300AM-Ert2 and Mef2cD/p300AM-Ert2 demonstrating specificity of Notch target sites in Hes1, Nrarp, and Cdh6, and the Mef2c targets Hdac9, Sgcg, and Cdkl5.