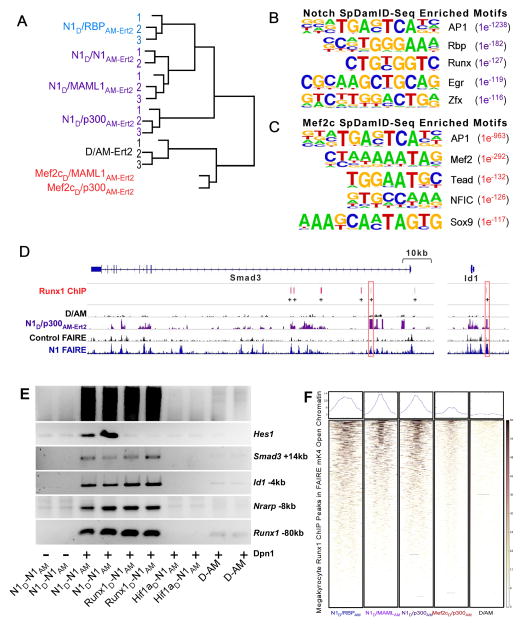
Figure 2. Genome-wide Analysis of SpDamID and Cooperative Labeling by Notch and Runx1 (see also Figure S2).
(A) Unsupervised hierarchical clustering of SpDamID-seq libraries. (B) The top 5 enriched TF motifs based on a HOMER analysis of N1D/p300AM-Ert2 SpDamID-seq. (C) The top 5 enriched motifs in the Mef2cD/p300AM-Ert2 SpDamID-seq library identified by Homer analysis. (D) Alignment of Runx1 ChIP in megakaryocytes with SpDamID and FAIRE identifies candidate enhancers with RBP and Runx1 binding within a 100bp window near the Smad3 and Id1 genes. (E) Biological duplicates of SpDamID libraries produced by Notch dimers and Runx1/Notch pairs but not by the Hif1a/Notch pair. The Notch dimers and Runx1/Notch pairs enriched for enhancers containing predicted binding sites for both factors within 100bp of each other (Smad3 +14kb, Id1 −4kb, Nrarp −8kb and Runx1 −80kb) but not for the Notch-only target (Hes1 −0.1kb). F) Heat maps of SpDamID and ChIP libraries centered on megakaryocyte Runx1 peaks present in FAIRE peaks from mK4 cells.