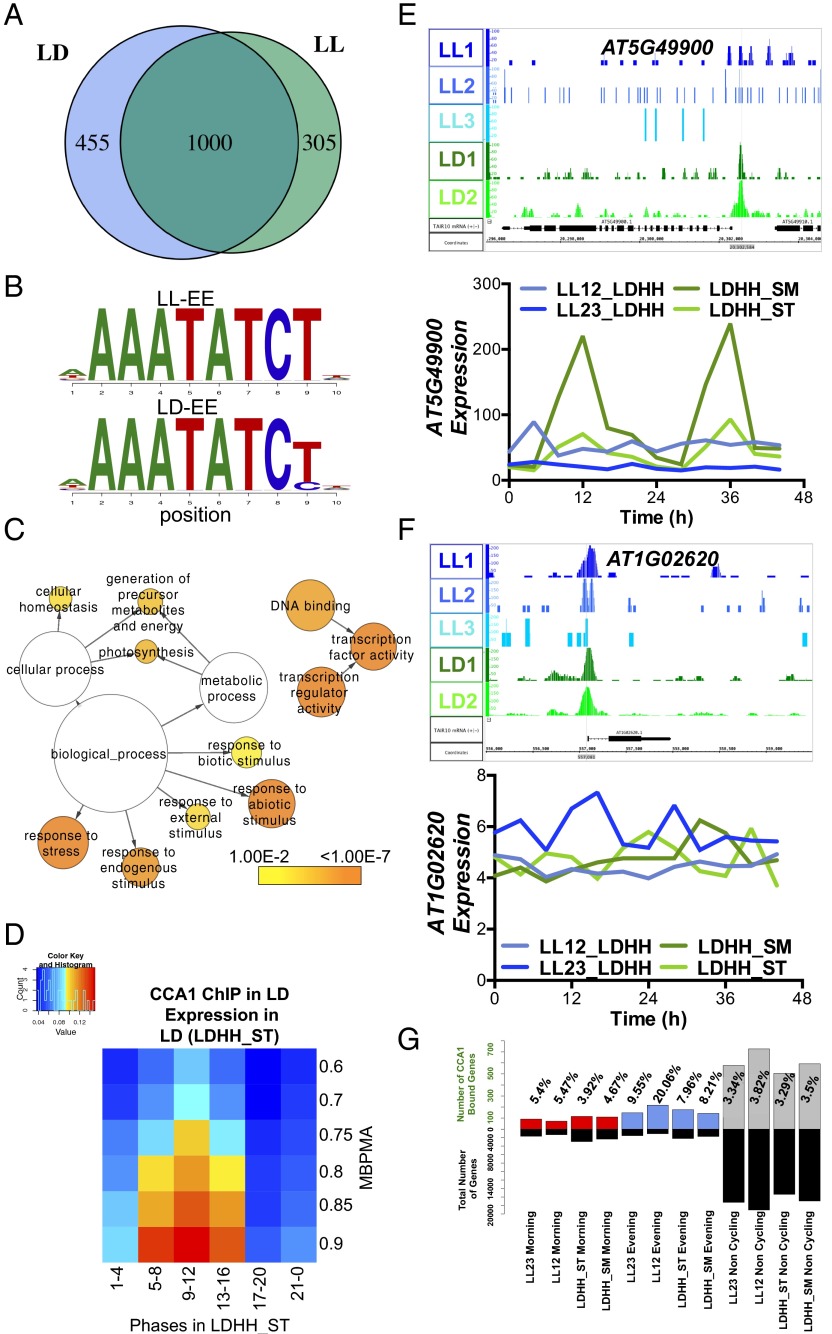
Fig. 2.
(A) Comparison of CCA1-occupied peaks in LL and LD. The Venn diagram shows the number of targets that are LD-specific (455) shaded in blue, the targets that are LL-specific (305) shaded in green, and the overlap between LD and LL targets (1,000 targets). (B) Alignment of the EE motif identified in LL and LD targets. (C) Functional enrichment analysis against GO Slim of the 1,231 CCA1 targets in LD with a peak within 1 kb of the TSS. The sizes of circles are based on the gene numbers. The color of each circle represents the enrichment P value for the GO term label on that circle, with orange representing highest enrichment and yellow the lowest enrichment above the cutoff (FDR corrected 0.01). (D) Phase enrichment of cycling CCA1-occupied targets. The expression datasets include transcripts that display >0.5 MBPMA in LD conditions [LDHH_ST, entrained in 12-h:12-h LD cycles and constant temperature (HH), data are from the Stitt laboratory (ST)] (39). Genes were grouped by phase and MBPMA score. The ratio of CCA1 occupancy to the number of genes in each group is indicated by intensity of the heat map (red and blue colors indicate maximum and minimum ratios of putative targets in the bin, respectively). (E) Example of a CCA1-occupied target in LD visualized using IGB (55). β-Glucosidase, GBA2 type family protein (AT5G49900), shows a rhythmic expression pattern in two of the previously published LD expression experiments (green) but not in LL (blue). (F) An example of a noncycling CCA1-occupied target in LD and LL, AT1G02620, visualized using IGB (55). Genome annotation for target gene structures is shown below. (G) Percentage of the CCA1-occupied targets in LD for each category of expression in each of the four time series datasets. The upper bars represent the percent of CCA1 targets that match the category: red bars represent morning-phased (ZT22–ZT4) genes, blue bars represent evening-phased (ZT10– ZT16) genes, and gray bars represent noncycling genes (MBPMA <0.80). The lower black bars show the total number of genes matching the respective category in that particular dataset.