Fig. 4.
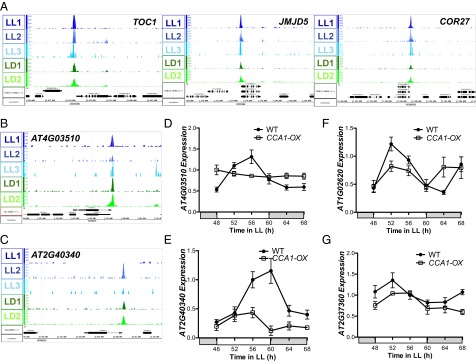
(A–C) CCA1-occupied peaks for the previously identified CCA1 targets TOC1 (AT5G61380), JMJD5 (AT3G20810), and COR27 (AT5G42900) (A) and uncharacterized CCA1 targets RMA1 (AT4G03510) (B) and DREB2C (AT2G40340) (C). Normalized tag counts by location in the genome are shown for each of the three replicate LL ChIP-Seq experiments (blue) and for each of the two replicate LD ChIP-Seq experiments (green). Genome annotation for each gene structure, visualized using IGB (55), is shown below. (D–G) qRT-PCR of wild-type and CCA1-OX plants grown in LL after 10-d entrainment in 12-h:12-h LD cycles for RMA1 (D), DREB2C (E), AT1G02620 (F), and ABCG2 (AT2G37360) (G). mRNA levels were normalized to IPP2 and PP2A expression (mean values ± SD, n = 2, two independent experiments).
