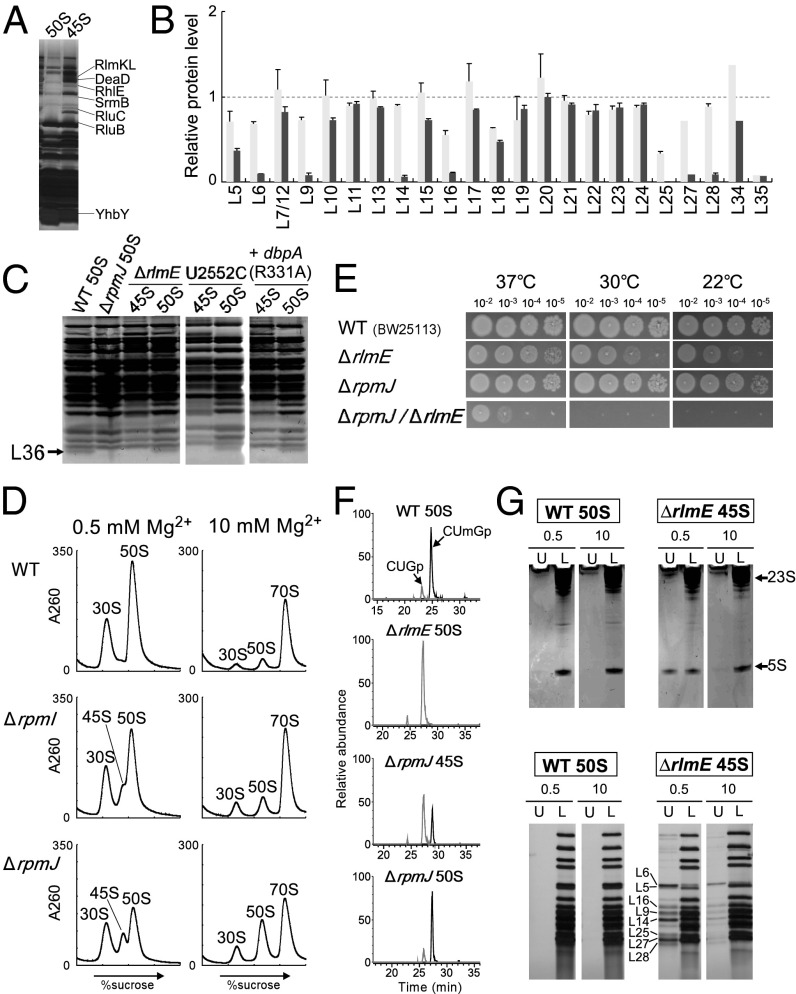
Fig. 4.
Involvement of L36 in RlmE-mediated late assembly. (A) Identification of assembly factors bound to the 45S precursor accumulated in ΔrlmE. Protein components of the 45S precursor were dissolved by SDS/PAGE. Each band was identified by peptide mass fingerprinting. (B) Relative level of ribosomal proteins in the 45S precursor versus WT 50S subunit treated (black bars) or untreated (gray bars) with high-salt solution. Relative levels of 23 ribosomal proteins were analyzed by the SILAC method. Tryptic peptides used for this analysis are listed in Table S1. The graph shows the averaged values with SD of the intensity ratios of three tryptic peptides for each protein. In the case of L27, L34, and L35 (Table S1), only one peptide was detected and available for this analysis. (C) Ribosomal proteins from the 45S precursor and the 50S subunit were resolved by urea/Tris-Tricine SDS 18% PAGE. The gel was stained with SYPRO Ruby Protein stain. Black arrow denotes L36. The 45S precursor and 50S subunit from ΔrlmE, the U2552C mutant, and a DbpA R331A-overexpressing strain were analyzed. WT and ΔrpmJ 50S subunits were used as controls. (D) SDG profiles of ribosomes from WT (Top), ΔrpmI (Middle), and ΔrpmJ (Bottom), performed at low Mg2+ concentration (Left) or high Mg2+ concentration (Right). All strains were cultured at 22 °C. (E) Severe growth defect of ΔrlmE/ΔrpmJ. Overnight saturated cultures of WT(BW25113) (Top), ΔrlmE (Second Panels), ΔrpmJ (Third Panels), and ΔrlmE/ΔrpmJ (Bottom) were serially diluted (102- to 105-fold dilutions), spotted onto LB plates, and cultivated at 37 °C for 18 h (Left), 30 °C for 24 h (Middle), or 22 °C for 51 h (Right). (F) Mass-spectrometric analysis of 2′-O-methylation at position 2552 in 23S rRNA from the indicated strains; WT 50S (Top), ΔrlmE 50S (Second), ΔrpmJ 45S (Third), and ΔrpmJ 50S (Bottom). The mass chromatograms represent CUmGp (m/z 987.13; black line) and CUGp (m/z 973.12; gray line) from the RNA segment G2529–C2579 of the 23S rRNA. (G) High-salt stripping analysis of the 45S precursor. (Upper) The result of denaturing 10% PAGE analyses of rRNAs from the 50S subunit and the 45S precursor in the upper fraction (U) and lower fraction (L), after high-salt stripping at low (0.5 mM) or high (10 mM) Mg2+ concentration. The gels were stained by ethidium bromide. (Lower) The result of 15% SDS/PAGE analyses of ribosomal proteins from the 50S subunit and 45S precursor in the upper fraction (U) and lower fraction (L), after high-salt stripping at low (0.5 mM) or high (10 mM) Mg2+ concentration. The gels were silver-stained.