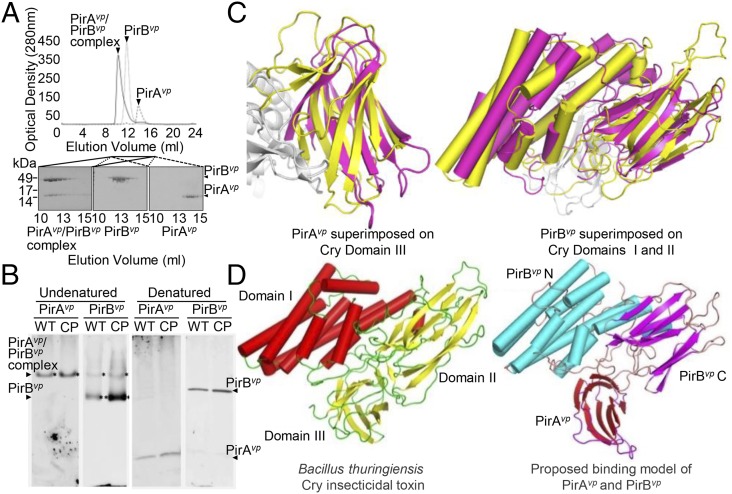
Fig. 5.
Formation of the PirABvp complex in vitro and in vivo, and a proposed binding model that suggests a functional similarity between PirABvp and the Cry proteins. (A) When purified PirAvp, PirBvp, and a PirAvp/PirBvp mixture were analyzed separately by using a Superose 16 gel filtration column (GE Healthcare), we found that, for the PirAvp/PirBvp mixture, both proteins were eluted faster than the individual proteins on their own. This suggests that PirAvp and PirBvp have formed a complex. (B) In the supernatant from a midlog-phase culture of the WT and the pVA1-cured+pirABvp (CP) strains, PirBvp was found in a complex with PirAvp and as a monomer, whereas PirAvp was only detected in the complex. Immunoblotting was performed with anti-PirAvp or anti-PirBvp antibody under undenatured, native PAGE conditions (Left). The denatured SDS/PAGE (β-mercaptoethanol; Right) confirmed that PirAvp and PirBvp were present in the culture medium. (C) Structural comparison of PirAvp and PirBvp with Cry. (Left) PirAvp (magenta) is superimposed on domain III of Cry (yellow). (Right) PirBvp (magenta) is compared with domains I and II of Cry. PirAvp has an rmsd of 3.2 Å for 88 matched Cα atoms in CRY domain III, and PirBvp superimposes with Cry domain I and II with an rmsd of 2.8 Å for 320 matched Cα atoms. (PDB ID codes: PirAvp, 3X0T; PirBvp, 3X0U). (D) A putative model of the PirABvp heterodimer was constructed by superimposing PirAvp on domain III of Cry and PirBvp on domains I and II. (Left) Cry toxin shown for comparison (PDB ID codes for Cry, 1CIY and ref. 30).