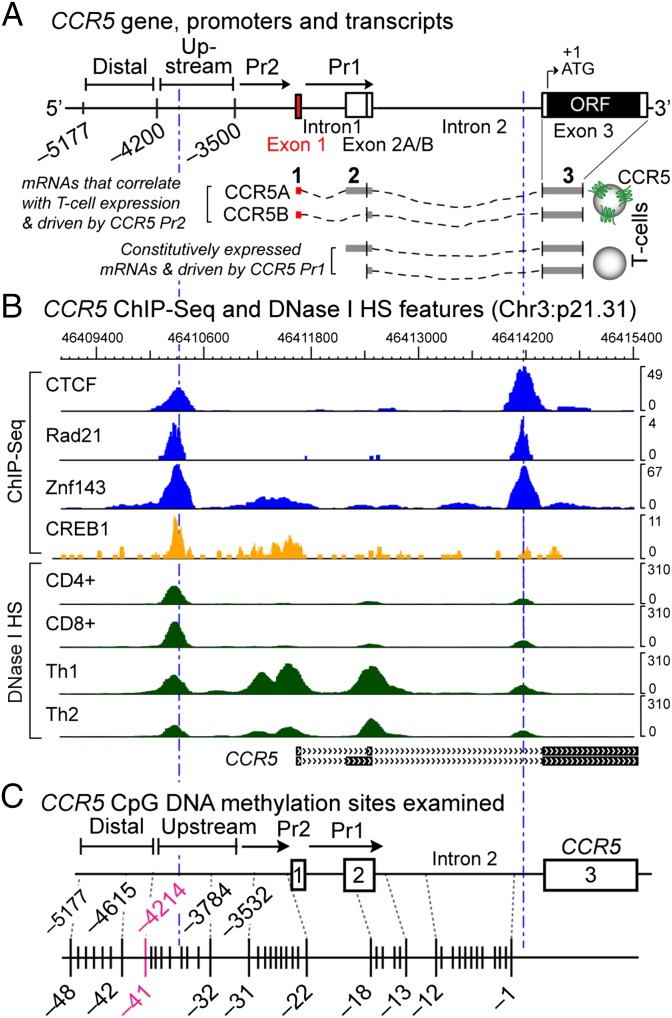
Fig. 1.
CCR5 gene, mRNA structure, and transcriptional and DNA methylation landmarks. (A) Three-exon CCR5 gene structure, two promoters, and exon 1-containing (full-length) vs. -lacking (truncated) mRNA isoforms (6). The upstream region starts ∼4.2 kb upstream of the ORF and indicates where increased micrococcal nuclease (Mnase) accessibility is apparent in memory cells compared with naïve cells (6). (B) Wiggle plots depict colocalization of CTCF, cohesin component Rad21, Znf143, and the transcription factor CREB1 (by ChIP-seq in GM12878 lymphoblastoid cells) and DNase I hypersensitivity sites in primary CD4+ and CD8+ T cells and Th1 and Th2 cells; derived from publicly available data. (C) CpG sites examined.