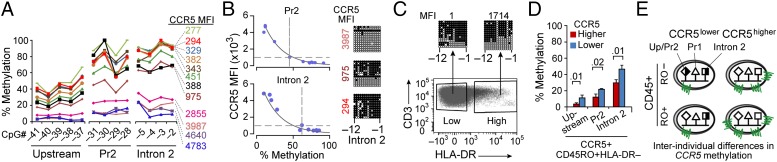
Fig. 4.
Methylation content of CCR5 cis-regulatory regions as a basis for interindividual differences in CCR5 levels on T cells. (A) Methylation content at the indicated CpGs in sorted CCR5+CD3+ T cells obtained from four healthy donors with the indicated postsort levels of CCR5 MFI (four gates per donor), assessed by pyrosequencing of representative sites. Lines connecting the data points are provided for better visualization of the data. (B) Relationship between methylation status of CCR5 cis-regions (promoter 2 and intron 2) and CCR5 levels (MFI). The line of best fit was determined by fitting the data to a generalized linear model using the exponential distribution. CCR5 methylation was calculated as average percent methylation in indicated cis-regions from pooled data from the same donors as in A. (Right) BGS validation of the inverse correlation between CCR5 expression and DNA methylation content of the indicated CpGs in CCR5-intron 2 derived from CCR5+ T cells with the indicated postsort MFI. (C) HLA-DR+ CD3+ T cells were sorted based on HLA-DR MFI using two arbitrarily set cell-sorting gates designated as low and high HLA-DR–expressing T cells. The methylation content in the sorted cells was determined by BGS. The numbers above the BGS data indicate HLA-DR+ MFI in postsorted T-cell fractions. (D) Methylation levels assessed by pyrosequencing of representative CpGs in the indicated CCR5 cis-regions in CCR5+CD45RO+HLA-DR− CD4+ T cells from three HIV− persons with higher and three persons with lower CCR5 expression (for results from CCR5+CD45RO+HLA-DR+ CD4+ T cells, see SI Appendix, Fig. S3A). The mean percentage methylation values are shown and the error bars depict SEM. (E) Model based on criterion 2.