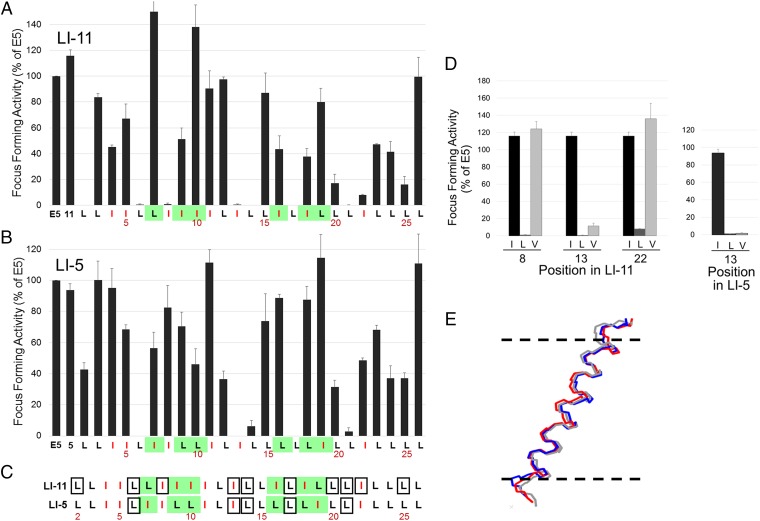
Fig. 3.
Comprehensive mutational analysis of traptamers. (A) C127 cells were infected with low-titer retrovirus expressing the E5 protein, wild-type LI-11, or an LI-11 mutant in which a leucine was changed to isoleucine or an isoleucine was changed to leucine. Sequence of wild-type LI-11 starting at position 2 is shown at the bottom, with isoleucines shown in red and positions that differ between LI-5 and LI-11 highlighted in green. The bars above the sequence show the activity of the mutant with the substitution at that position. Transformed foci were counted after infected cells were incubated at confluence for 3 wk. Focus-forming activity displayed as in Fig. 1E. (B) Focus-forming activity of LI-5 mutants tested and displayed as in A. (C) Sequences of LI-5 and LI-11 are displayed as described in A. Residues that when mutated to leucine or isoleucine result in 80% or greater reduction in focus-forming activity are shown in boxes. (D) Focus-forming activity was measured and displayed as described in A for traptamers containing isoleucine (black), leucine (dark gray), or valine (light gray) at the indicated position in traptamer LI-11 (Left) or at position 13 in LI-5 (Right). (E) Molecular dynamics simulation indicates that the helical backbones of LI-11, LI-11-I13L, and LI-11-I13V are very similar. The diagram shows an overlay of lateral views of the backbones of three aligned helices. The horizontal dashed lines represent the approximate extent of the membrane bilayer hydrophobic region; the amino termini of the proteins are at the top.