Abstract
Lung cancer is the leading cause of cancer death worldwide. Although several genetic variants associated with lung cancer have been identified in the past, stringent selection criteria of genome-wide association studies (GWAS) can lead to missed variants. The objective of this study was to uncover missed variants by using the known association between lung cancer and first-degree family history of lung cancer to enrich the variant prioritization for lung cancer susceptibility regions. In this two-stage GWAS study, we first selected a list of variants associated with both lung cancer and family history of lung cancer in four GWAS (3,953 cases, 4,730 controls), then replicated our findings for 30 variants in a meta-analysis of four additional studies (7,510 cases, 7,476 controls). The top ranked genetic variant rs12415204 in chr10q23.33 encoding FFAR4 in the Discovery set was validated in the Replication set with an overall OR of 1.09 (95% CI = 1.04, 1.14, P = 1.63 × 10−4). When combining the two stages of the study, the strongest association was found in rs1158970 at Ch4p15.2 encoding KCNIP4 with an OR of 0.89 (95% CI = 0.85, 0.94, P = 9.64 × 10−6). We performed a stratified analysis of rs12415204 and rs1158970 across all eight studies by age, gender, smoking status, and histology, and found consistent results across strata. Four of the 30 replicated variants act as expression quantitative trait loci (eQTL) sites in 1,111 nontumor lung tissues and meet the genome-wide 10% FDR threshold.
Keywords: lung cancer, family history, secondary phenotype, genetic susceptibility, genome-wide association studies, eQTL
Introduction
Lung cancer is the leading cause of cancer death worldwide [Ferlay et al., 2010]. Genome-wide association studies (GWAS) have identified several genetic susceptibility loci for lung cancer including ch15q25, 5p15, 6p21, 9p21, and 12p13 [Amos et al., 2008; Hung et al., 2008; McKay et al., 2008; Shi et al., 2012; Timofeeva et al., 2012; Wang et al., 2008, 2014]. However, these loci only accounted for a small fraction of the heritability of lung cancer. A main issue in understanding the genetic architecture of complex diseases like lung cancer is rooted in the standard analytical approach currently used for GWAS, which typically involves identical statistical tests for each single marker and conservative multiple testing corrections to account for the large number of variants investigated in GWAS. Consequently, many genetic variants affecting lung cancer risk have gone undiscovered.
One way to unveil these hidden variants is to take additional phenotypes linked to lung cancer risk into consideration during the statistical testing process. Performing a secondary phenotype analysis in addition to the primary phenotype analysis allows for increased use of the information collected within each study. Secondary phenotype analysis may be used to gather information about variants potentially associated with the primary phenotype, for instance by reducing the number of variants investigated in the primary phenotype analysis or by giving priority to variants that are also consistently associated with the secondary phenotype.
For the genetics of lung cancer, a natural choice of secondary phenotype is family history of lung cancer among first-degree relatives, since it has been shown to be associated with lung cancer risk and it would reflect the genetic component of lung cancer etiology [Cote et al., 2012]. The genetic variants associated with family history of lung cancer would be considered to have a higher probability of being linked to lung cancer risk. In other words, all else being equal, the variants associated with both family history of lung cancer and lung cancer risk would have a higher probability of representing a true association compared to those associated with lung cancer risk alone.
A number of procedures have been proposed for the analysis of secondary phenotypes in case–control studies, such as the inverse-probability-of-sampling-weighted (IPW) regression and a likelihood-based method that takes into account the case–control sampling of the data [Lin and Zeng 2009; Monsees et al., 2009; Richardson 2007]. With IPW regression, the study base is approximated by upweighting each sampled subject such that he/she represents multiple subjects in the study base. This procedure has been shown to avoid the bias in regression estimates that is related to the nonrandom sampling of cases and controls from the population, but with the cost of reduced power [Monsees et al., 2009]. Alternatively, Lin and Zeng’s likelihood-based analysis for secondary phenotypes provides maximum likelihood estimates obtained by reflecting the case–control sampling in the analysis. In our analysis, we have chosen to use the method proposed by Lin and Zeng, because the resulting estimates are asymptotically unbiased, the procedure is both statistically and computationally efficient, and the online software offers ease of computation.
To enrich the GWAS analysis with a secondary phenotype, we conducted a two-stage analysis to uncover additional genetic loci for lung cancer risk, focusing on those that did not reach GWAS significance. In the first stage, we prioritized the variants based on their association with lung cancer risk, with higher prior probability assigned to those that were also associated with family history of lung cancer. In the second stage, we validated prioritized variants using an independent National Cancer Institute (NCI) lung cancer GWAS dataset from the database for Genotypes and Phenotypes (dbGaP) [Landi et al., 2009] and three datasets newly genotyped with an Axiom array. Finally, the functional meaning of the newly identified lung cancer SNPs were extended to gene expression levels in human lung tissues.
Subjects and Methods
Stage 1
Four lung cancer GWAS in the International Lung Cancer Consortium were used to identify variants associated with both lung cancer and first-degree family history of lung cancer: the Toronto study [Hung et al., 2008; McKay et al., 2008], the Central Europe study [Hung et al., 2008], the MD Anderson study [Amos et al., 2008], and the Germany study [Landi et al., 2009; Sauter et al., 2008]. A total of 3,953 lung cancer cases and 4,730 controls were included in the Discovery set from the four studies, and the study characteristics of these four studies are summarized in Table 1. Unconditional logistic regression was used to assess the association between genetic variants and lung cancer risk, adjusted by age, sex, ever/never smoking (except in the MD Anderson study as all subjects were smokers) and center (for the Central Europe study) using PLINK software [Purcell et al., 2007]. The associations across all four studies were then summarized using a fixed effects logistic regression model with GWAMA software [Magi and Morris, 2010]. Variants that were present in at least two studies were included in the Discovery meta-analysis.
Table 1.
Individual study characteristics among Caucasians according to primary phenotype (lung cancer)
| Stage | Study | Location | Study period | Study design | Case no. | Control no. | Genotyping platform |
|---|---|---|---|---|---|---|---|
| Discovery (3,953 cases and 4,730 controls) |
Toronto | GTA, Canada | 1997–2002 | Population and Hospital CC | 331 | 499 | Illumina HumanHap300 |
| IARC | Central Europe | 1998–2002 | Hospital CC | 1,964 | 2,610 | Illumina HumanHap300 | |
| MDACC | Texas, USA | 1997–2007 | Hospital CC | 1,154 | 1,137 | Illumina HumanHap300 | |
| HMGU | Germany | 2000–2008 (LUCY) | Population CC | 504 | 484 | Illumina HumanHap550 | |
| 1990–1998 (KORA) | |||||||
| 1997–2007 (Heidelberg) | |||||||
| Replication (7,510 cases and 7,476 controls) |
NCI (dbGaP) | USA and Italy | 1985–2005 | Population CC | 5,699 | 5,815 | HumanHap550v3.0 |
| Human610_Quadv_1_B | |||||||
| Human1M_Duov3_b | |||||||
| HumanHap300v1.1 | |||||||
| HumanHap250Sv1.0 | |||||||
| MSH-PMH | Toronto, Canada | 2009–2013 | Clinic CC | 1,073 | 939 | Axiom array | |
| MEC | USA | 1993–1996 | Nested CC | 215 | 225 | Axiom array | |
| Harvard | USA | 1990–2006 | Nested CC | 523 | 497 | Axiom array |
To identify variants associated with first-degree family history of lung cancer we utilized a likelihood-based approach [Lin and Zeng, 2009] that takes into account the lung cancer case–control status. A standard analysis of a secondary phenotype from a case–control study using an unconditional logistic regression model may be biased if the primary and secondary phenotypes are associated with one another and if the variant under study is associated with each phenotype [Monsees et al., 2009]. For a dichotomous secondary phenotype, Lin and Zeng [2009] use the logistic regression model,
| (1) |
where Y denotes the secondary phenotype, X denotes the genotype score for some SNP, and we are interested in solving for β1. Furthermore, the secondary phenotype and the genotype are related to the primary phenotype (D) by
| (2) |
By inserting equations (1) and (2) into the retrospective likelihood,
| (3) |
where P (Di = 1) = Σy Σx P (Di = 1|x, y) P (y|x) P (x), P (Di = 0) = 1 − P (Di = 1), and P (Di = 0|XiYi) = 1 − P (Di = 1|XiYi). The Newton–Raphson algorithm may be used to maximize Equation (3) and inferences may be made on β1 using a Wald or a likelihood ratio statistic. Lin and Zeng [2009] provide a computationally efficient online software program called SPREG to implement this procedure.
To systematically incorporate the secondary phenotype association into the analysis of the primary phenotype, we used the Bayesian false discovery probability (BFDP) [Wakefield, 2007, 2009]. Briefly, the BFDP estimates the false Discovery probability (or the probability of the null hypothesis given the data) to identify noteworthy associations. The BFDP was obtained for the association between variants and lung cancer by setting the on the null to prior = (1-p2) (weight), where p2 represents the secondary phenotype P value and the weight is set to the proportion of the four studies having the secondary phenotype ORs in the same direction. Thus, the greater the evidence of association between variant and family history, the greater was the prior odds on the null. The selected variants were sorted by the BFDP obtained using the primary and secondary phenotype results of the Discovery analysis.
Variants with associations at the 5% significance level with both lung cancer and family history of lung cancer in the same direction in each meta-analysis or variants highly associated with family history of lung cancer with P values s 0.0001 in the meta-analysis of the secondary phenotype were selected to be validated. A total of 537 variants were selected for replication.
In addition, we further selected 215 of the 537 variants to be included in the custom panel of Axiom array for fine mapping and validation purposes. This selection was based on consistent direction of lung cancer odds ratios between the Discovery set of four studies (3,953 cases and 4,730 controls), the pilot Replication set based on an independent dbGaP lung cancer study (2,783 cases and 2,713 controls, accession no. pht000119) [Landi et al., 2008], and the combined P values less than 0.05 based on the meta-analysis of all five studies. Of the 215 nominated variants, 203 were successfully included in the custom panel of the Axiom array after the design and quality control stage.
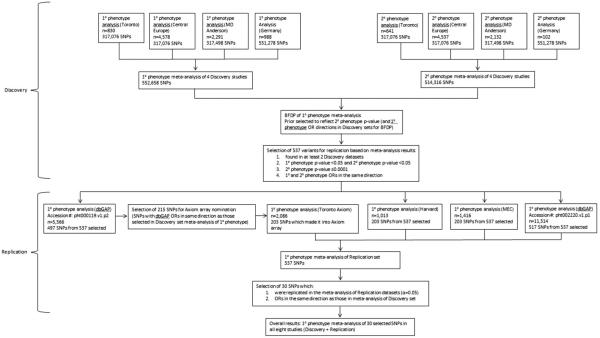
A flow chart of the selection of variants in the Discovery and Replication sets is included in Figure 1.
Figure 1.
Flow chart of Discovery and Replication study design. The Discovery stage includes primary (lung cancer) and secondary (family history of lung cancer) phenotype analysis of four GWASs, with a meta-analysis for each phenotype. The Replication stage includes four additional studies which were used to validate associations between SNPs and lung cancer.
Stage 2
The final Replication stage consisted of four studies: a large dbGaP lung cancer study (accession no. pht02220.v1.p1, an expansion of our pilot Replication set based on NCI dbGaP study accession no. pht000119) using in silico look up [Landi et al., 2009], and a custom Axiom array with variants nominated from our Discovery analysis genotyped in three studies: lung cancer case–control study conducted in Mount Sinai Hospital and Princess Margaret Hospital in Toronto (MSH-PMH study) [Wang et al., 2014], Nurses’ Health Study at Brigham and Woman’s Hospital and Harvard Medical School [Colditz et al., 2007], and the Multi-Ethnic Cohort (MEC) study [Derby et al., 2008]. A total of 7,510 lung cancer cases and 7,476 controls were included in the Replication set based on the four studies, and the study characteristics are summarized in Table 1.
The in silico validation dataset from NCI study (dbGAP accession #: pht002220.v1.p1) contained 517 of the 537 variants selected for validation, while the Axiom custom panel genotyped in the Toronto, Harvard, and MEC studies contained 203 of the variants. All of the 537 variants were available from either the NCI in silico look up and/or the Axiom custom panel. An unconditional logistic regression model, adjusted for age, sex, and smoking status was used to detect variants associated with lung cancer in each Replication dataset. Note that the NCI dbGAP dataset was adjusted for age and sex only due to a lack of smoking information.
The results of the four Replication studies were summarized using a fixed effects meta-analysis. The variants were considered validated if they (i) were statistically significant at the 5% level in the Replication meta-analysis, and (ii) had effect estimates in the same direction as those in the Discovery analysis.
We also conducted stratified analysis by age of onset, smoking status, and histology for the top variants of interests to assess the potential differential effects. The stratified analysis of histology and smoking status were adjusted for age and sex. This analysis was performed in each of the eight datasets investigated, and the results were summarized using fixed-effects meta-analysis in GWAMA [Magi and Morris, 2010].
Finally, to gain better insight into the molecular mechanisms underlying these associations, we were interested in finding whether selected variants act as eQTL sites in lung tissue. We had access to a lung-specific eQTL study which reported cis- (within 1 Mb of transcript) and trans- (further than 1 Mb or on a different chromosome) acting eQTL. The study is a meta-analysis of nontumor lung tissue eQTL data from Laval University, University of British-Columbia, and University of Groningen based on 1,111 patients who underwent resectional surgery. Gene expression profiles were obtained using an Affymetrix array testing 51,627 noncontrol probesets, and DNA genotyping was performed using the Illumina-Human 1M-Duo BeadChip array. A robust linear model adjusted for age, gender, and smoking status was implemented to find associations between genetic variants and gene expression. The eQTLs identified at 10% false discovery rate (FDR) in each site as well as in the meta-analysis of all sites were then reported. Further details of the methods and analysis have previously been reported [Hao et al., 2012; Lamontagne et al., 2013; Obeidat et al., 2013; Thun et al., 2013; Wain et al., 2014].
Results
Stage 1
A total of 317,924 variants were assayed in at least two Discovery datasets and were included in the Discovery meta-analysis. Among these variants, 537 variants were shown to be associated with both lung cancer risk and having a family history of lung cancer among first-degree relatives at P values < 0.05 with consistent direction of ORs. These 537 variants were selected to move forward into the Replication set.
Stage 2
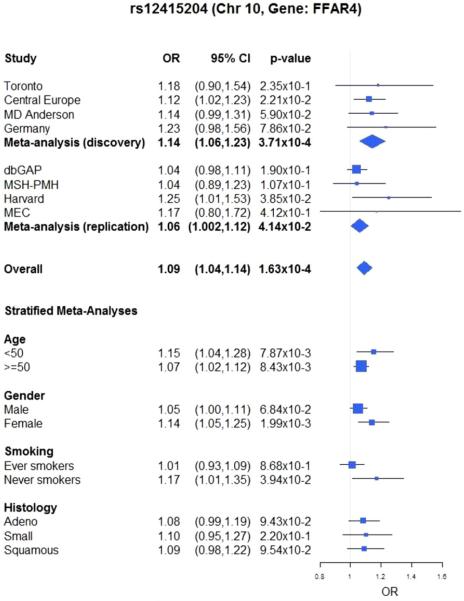
Thirty of the 537 variants selected in Stage 1 were found to be statistically significantly associated with lung cancer at the 5% level in the replication analysis and had consistent effect direction with the Discovery analysis. Table 2 presents meta-analysis results for the association of the 30 variants. When ranked by BFDP, the evidence was strongest for ch10q23.33 encoding the gene FFAR4 marked by sequence variant rs12415204, which is a nucleotide change from C to A with minor allele frequency ranging from 21% to 25% across all studies. The A allele was positively associated with family history of lung cancer with an OR of 1.18 (95% CI = (1.02, 1.37), P = 2.55 × 10−2) and had an association with increased risk of lung cancer with an OR of 1.09 (95% CI = (1.04, 1.14), P = 1.63 × 10−4) in the Discovery set, with a BFDP of 0.047. It was replicated in the Replication datasets with an OR of 1.06 (95% CI = (1.00, 1.12), P = 4.14 × 10−2).
Table 2.
Variants which were associated with lung cancer and first degree family history of lung cancer and were validated in Stage 2
| Ref. effect allele |
Position (NCBI Build 36) |
Meta-analysis (Discovery) |
Meta-analysis (Replication) |
Combined-analysis (overall) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Gene | SNP | OR (FH) | P value (FH) | OR (LC) | P value (LC) | BFDP (LC) | OR (LC) | P value (LC) | OR (LC) | p-value (LC) | ||
| 1 | FAM5C | rs6678713 | A,G | 188,656,364 | 1.17 | 0.0236 | 1.08 | 0.0270 | 0.7180 | 1.06 | 0.0360 | 1.06 | 0.0026 |
| 2 | ALLC | rs6542651 | T,C | 3,737,705 | 1.26 | 0.0443 | 1.17 | 0.0070 | 0.7913 | 1.10 | 0.0219 | 1.13 | 0.0006 |
| 2 | DPP10 | rs9308697 | T,C | 115,124,129 | 1.19 | 0.0254 | 1.09 | 0.0191 | 0.6270 | 1.08 | 0.0072 | 1.08 | 0.0006 |
| 4 | KCNIP4 | rs1158970 | C,T | 20475567 | 0.78 | 0.0136 | 0.90 | 0.0130 | 0.9657 | 0.89 | 0.0002 | 0.89 | 9.64E-06 |
| 4 | FRAS1 | rs1546318 | T,G | 79,168,396 | 1.25 | 0.0283 | 1.16 | 0.0053 | 0.2854 | 1.11 | 0.0187 | 1.13 | 0.0003 |
| 4 | FRAS1 | rs1393644 | A,C | 791,757,31 | 1.25 | 0.0310 | 1.15 | 0.0072 | 0.3474 | 1.09 | 0.0190 | 1.11 | 0.0008 |
| 6 | ATXN1 | rs17669356 | T,C | 16,801,065 | 0.80 | 0.0154 | 0.92 | 0.0344 | 0.7315 | 0.94 | 0.0377 | 0.93 | 0.0033 |
| 6 | C6orf205 | rs2844680 | G,T | 31054475 | 1.16 | 0.0294 | 1.10 | 0.0498 | 0.5168 | 1.07 | 0.0055 | 1.07 | 0.0009 |
| 6 | PKIB | rs7452823 | T,C | 123,047,594 | 0.87 | 0.0484 | 0.94 | 0.0486 | 0.8028 | 0.93 | 0.0031 | 0.94 | 0.0007 |
| 7 | TSPAN13 | rs2389700 | C,T | 16,788,162 | 1.27 | 0.0066 | 1.11 | 0.0164 | 0.5756 | 1.08 | 0.0398 | 1.09 | 0.0019 |
| 7 | MTERF | rs2157998 | T,C | 91158790 | 0.84 | 0.0187 | 0.93 | 0.0343 | 0.9834 | 0.95 | 0.0499 | 0.95 | 0.0062 |
| 8 | ZFPM2 | rs285823 | T,C | 106,171,356 | 1.17 | 0.0264 | 1.09 | 0.0128 | 0.5605 | 1.07 | 0.0102 | 1.08 | 0.0002 |
| 8 | ADCY8 | rs7017572 | G,A | 132,398,482 | 1.14 | 0.0438 | 1.07 | 0.0416 | 0.5655 | 1.07 | 0.0191 | 1.07 | 0.0019 |
| 9 | PTPRD | rs10959312 | C,T | 10,802,417 | 0.80 | 0.0116 | 0.89 | 0.0043 | 0.9330 | 0.94 | 0.0343 | 0.92 | 0.0004 |
| 9 | DBH | rs2007153 | C,T | 135,493,640 | 0.87 | 0.0499 | 0.92 | 0.0175 | 0.9258 | 0.95 | 0.0280 | 0.94 | 0.0012 |
| 10 | CACNB2 | rs7901587 | C,T | 18,447,963 | 1.27 | 0.0071 | 1.10 | 0.0432 | 0.9932 | 1.08 | 0.0252 | 1.09 | 0.0026 |
| 10 | FFAR4 | rs12415204 | C,A | 95,320,880 | 1.18 | 0.0255 | 1.14 | 0.0004 | 0.0471 | 1.06 | 0.0414 | 1.09 | 0.0002 |
| 10 | SORCS1 | rs10509832 | C,T | 109,070,424 | 1.15 | 0.0426 | 1.07 | 0.0454 | 0.7953 | 1.08 | 0.0056 | 1.07 | 0.0007 |
| 11 | FAM76B | rs10831422 | T,C | 95,118,616 | 0.85 | 0.0269 | 0.92 | 0.0189 | 0.9577 | 0.94 | 0.0174 | 0.93 | 0.0010 |
| 11 | FAM76B | rs11021302 | G,A | 95,125,416 | 0.84 | 0.0153 | 0.93 | 0.0227 | 0.9812 | 0.94 | 0.0086 | 0.94 | 0.0013 |
| 11 | SORL1 | rs1503415 | A,C | 120,991,762 | 1.16 | 0.0368 | 1.07 | 0.0468 | 0.7932 | 1.06 | 0.0337 | 1.07 | 0.0037 |
| 13 | PCDH17 | rs7991911 | G,A | 58,036,178 | 1.25 | 0.0422 | 1.12 | 0.0497 | 0.7101 | 1.11 | 0.0085 | 1.12 | 0.0010 |
| 15 | CYP19A1 | rs10519295 | T,C | 49,319,939 | 0.79 | 0.0463 | 0.89 | 0.0402 | 0.9445 | 0.90 | 0.0124 | 0.91 | 0.0028 |
| 17 | VAMP2 | rs2278637 | T,G | 8,002,827 | 0.85 | 0.0257 | 0.93 | 0.0486 | 0.5776 | 0.94 | 0.0214 | 0.94 | 0.0025 |
| 18 | DYM | rs833520 | C,T | 44,818,557 | 1.24 | 0.0053 | 1.08 | 0.0395 | 0.9954 | 1.08 | 0.0211 | 1.08 | 0.0020 |
| 18 | DYM | rs10520770 | T,C | 44,856,962 | 0.86 | 0.0228 | 0.92 | 0.0072 | 0.4606 | 0.95 | 0.0283 | 0.94 | 0.0012 |
| 18 | DYM | rs2078286 | G,A | 45,132,860 | 0.85 | 0.0190 | 0.93 | 0.0317 | 0.9826 | 0.93 | 0.0107 | 0.93 | 0.0009 |
| 18 | CBLN2 | rs1443321 | G,A | 68,035,323 | 1.24 | 0.0183 | 1.1 | 0.0438 | 0.9819 | 1.08 | 0.0383 | 1.09 | 0.0040 |
| 19 | ZNF537 | rs2032868 | T,C | 36,692,866 | 0.83 | 0.0065 | 0.90 | 0.0300 | 0.6794 | 0.94 | 0.0216 | 0.93 | 0.0022 |
| 21 | C21orf34 | rs1014602 | G,T | 17,263,238 | 1.16 | 0.0483 | 1.14 | 0.0228 | 0.9088 | 1.07 | 0.0196 | 1.08 | 0.0014 |
Combining the four studies in the Discovery set and four studies in the Replication set for the 30 validated variants, rs12415204 remained one of the top hits (OR = 1.09, P = 1.63 × 10−4). When stratified by age, smoking status, and histology (Fig. 2), we observed nominally significant associations in both age groups, in females, and in never smokers. Furthermore, although the differences in ORs between strata for age, smoking status, and histology were not statistically significant (P > 0.05), patients with young onset (<50 years old), females, and never smokers had more prominent associations between lung cancer and rs12415204, a variant in FFAR4.
Figure 2.
Forest plot of the association between rs12415204 with lung cancer across eight studies, stratified by age, gender, smoking, and histology.
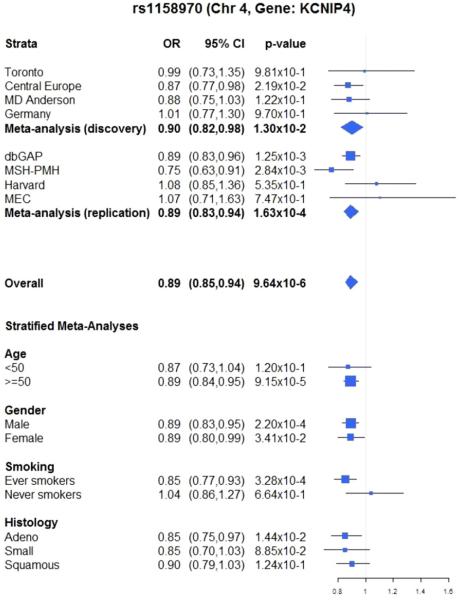
All of the replicated variants in Table 2 would have been overlooked in the Discovery stage had we focused solely on statistically significant associations with lung cancer at the genome-wide level (P < 10−8) and not taken into consideration the secondary phenotype meta-analysis results. For instance, the strongest signal in the Replication stage was observed for ch4p15, which encodes KCNIP4, with an OR of 0.89 for rs1158970 (P = 1.63 × 10−4). This variant had an OR of 0.90 (P = 1.30 × 10−2) for lung cancer and an OR of 0.78 (P = 1.36 × 10−2) for first-degree family history of lung cancer in the Discovery analysis. When combining eight studies from both stages, the strongest observed signal of the top 30 variants had an OR of 0.89 (95% CI = 0.85, 0.94, P value = 9.64 × 10−6) for lung cancer. Figure 3 displays the results of the association between rs1158970 and lung cancer in all eight studies, including a stratified analysis by age, gender, smoking, and histology. We observed consistent results across studies and across strata.
Figure 3.
Forest plot of the association between rs1158970 with lung cancer across eight studies, stratified by age, gender, smoking, and histology.
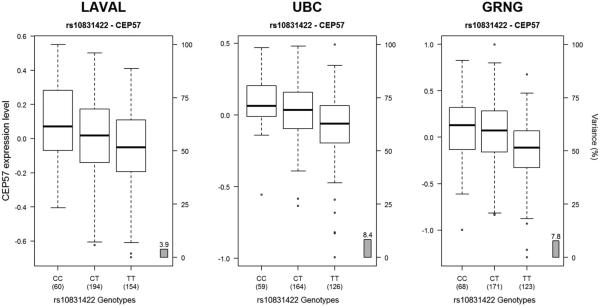
The replicated variants in Table 2 were then investigated to determine if they act as lung eQTLs. Table 3 presents the cis- and trans-eQTL analysis results for variants with at most 10% FDR. Four of the thirty variants were found to be lung eQTLs, with the strongest signals coming from two variants on chromosome 11 (rs10831422 and rs11021302) that were associated with the expression of CEP57 (P = 1.61 × 10−19) (Fig. 4).
Table 3.
Variants acting as lung eQTLs and meeting genome-wide significance of 10% FDR
| eQTL- regulated gene |
eQTL- regulated probeSet |
eQTL Z-statistic |
eQTL meta P value |
Lung cancer overall result OR, (95 CI%), P value |
|||
|---|---|---|---|---|---|---|---|
| eQTL | Laval | Groningen | UBC | ||||
| rs2844680 | LST1 | 100149313_TGI_at | 2.87 | 2.60 | 2.11 | 1.04 × 10–5 | 1.07, (1.03, 1.12), 0.000855 |
| G;T | CDSN | 100303039_TGI_at | −3.60 | −3.14 | −4.29 | 3.18 × 10–10 | |
| ZFP57 | 100303994_TGI_at | 1.46 | 6.20 | 3.51 | 1.21 × 10–10 | ||
| 100309580_TGI_at | 2.83 | 5.15 | 3.24 | 8.29 × 10–11 | |||
| rs10831422 | 100126872_TGI_at | −3.43 | −1.73 | −3.65 | 3.37 × 10–7 | 0.93, (0.89, 0.97), 0.000976 | |
| T;C | FAM76B | 100129097_TGI_at | −4.48 | −3.81 | −4.44 | 2.22 × 10–13 | |
| FAM76B | 100141148_TGI_at | −3.48 | −4.67 | −6.04 | 6.12 × 10–16 | ||
| CEP57 | 100127730_TGI_at | −4.39 | −5.52 | −5.81 | 1.61 × 10–19 | ||
| FAM76B | 100159174_TGI_at | 5.31 | 0.85 | 3.08 | 3.90 × 10–8 | ||
| rs11021302 | 100126872_TGI_at | −3.43 | −1.73 | −3.69 | 3.03 × 10–7 | 0.94, (0.90, 0.98), 0.00127 | |
| G;A | CEP57 | 100127730_TGI_at | −4.39 | −5.52 | −5.81 | 1.61 × 10–19 | |
| FAM76B | 100141148_TGI_at | −3.48 | −4.67 | −6.08 | 5.46 × 10–16 | ||
| FAM76B | 100129097_TGI_at | −4.48 | −3.81 | −4.49 | 1.84 × 10–13 | ||
| FAM76B | 100159174_TGI_at | 5.31 | 0.85 | 3.08 | 3.82 × 10–8 | ||
| rs2278637 | VAMP2 | 100137040_TGI_at | 3.85 | 2.50 | 5.52 | 7.10 × 10–12 | 0.94, (0.90, 0.98), 0.002478 |
| T;G | 100152407_TGI_at | 2.88 | 2.89 | 2.03 | 6.22 × 10–6 | ||
| TMEM107 | 100301520_TGI_at | 3.78 | 2.58 | 4.78 | 1.07 × 10–10 | ||
Note that in the final column, the final two SNPs have different P values, yet the same ORs and CIs. The similarity in effect size is due to rounding. The eQTL test statistics are based on the major allele, as defined under each rs number in column 1 (major allele; minor allele).
Figure 4.
Boxplots of gene expression levels in the lung for CEP57 according to genotype groups for SNP rs10831422. The left y-axis shows the mRNA expression levels for CEP57. The x-axis represents the three genotyped groups for SNP rs10831422. The right y-axis shows the proportion of the gene expression variance explained by the SNP (gray bar). Each panel represents a different cohort: Laval (n = 408), UBC (n = 349), Groningen (n = 362). The eQTL P values were 6.2 × 10−05, 4.9 × 10−07, and 1.7 × 10−07, respectively.
Discussion
In this analysis, we used the family history of lung cancer to enrich the analysis of lung cancer risk based on secondary phenotype regression and we identified 30 variants representing 25 independent loci, which were replicated in the two-stage analysis of eight studies with a total of 11,463 cases and 12,206 controls. The observed consistency and replication in the direction of the signals and in the strength of the signals across phenotypes provided increased evidence of true associations for a highly selected list of variants. The top ranked variant in the gene FFAR4 (rs12415204, ch10q23) was validated in the Replication set and it was ranked first by BFDP and second by overall P value after KCNIP4.
Classified as a fatty acid metabolism gene and previously named GPR120, FFAR4 has been shown to be associated with body mass index [Ichimura et al., 2012]. Although the expression of this gene has been identified as a tumor-promoting receptor in colorectal carcinoma [Wu et al., 2013], to our knowledge this gene has not previously been implicated in lung cancer carcinogenesis. Furthermore, our stratified analysis suggested a potentially greater effect among those with young onset (<50 years old) and those who never smoked. These observations are consistent with our expectations of the role of the genetic component in lung cancer etiology in these lower risk populations, which lends further evidence to a causal signal.
Although the difference in ORs between strata within each category was not found to be statistically significant, confidence intervals suggested the potential for biologically relevant associations by strata, particularly among patients with young onset (<50 years old), females, and never smokers. The nonsignificant test for heterogeneity may be a consequence of low power for this test. Thus, further study is recommended to understand these potential differences between strata. Larger studies may also conduct discovery analyses among these strata and identify other genetic variants that may only have a signal in populations with increased genetic susceptibility.
The KCNIP4 gene encodes small calcium-binding proteins and is part of the family of voltage-gated potassium channel-interacting proteins [Burgoyne, 2007]. This gene was proposed to be a candidate gene for renal cell carcinoma [Boone et al., 2007] and asthma [Himes et al., 2013]. It was recently identified as a lung cancer susceptibility gene when using the biological function of the gene and functional significance of the variants as the prior weighting in hierarchical modeling [Brenner et al., unpubl. ms]. The fact that KCNIP4 has been identified as a lung cancer susceptibility locus by two completely different approaches adds weight to the overall evidence of an association.
Both of these two top ranked variants would have been missed had they not been preselected using secondary phenotype association results. In fact, all of the 30 validated variants would have been missed at the genome-wide level of significance by ignoring secondary phenotype information.
Interestingly, a number of lung cancer associated variants were found to be lung eQTLs, suggesting that their effect on lung cancer risk is probably mediated through changes in gene expression, i.e., have regulatory function. The strongest eQTLs were for centrosomal protein 57kDa (CEP57) and family with sequence similarity 76, member B (FAM76B); two neighboring genes on chromosome 11. CEP57 encodes a protein called translokin, and is involved in the centrosomal localization and microtubular stabilization [Momotani et al., 2008], and in the trafficking of factors, such as fibroblast growth factor 2 (FGF2) [Meunier et al., 2009]. Mutations in CEP57 have been found to cause mosaic variegated aneuploidy syndrome [Snape et al., 2011]; a rare autosomal recessive disorder characterized by mosaic aneuploidies (a condition in which a person has one or a few chromosomes above or below the normal chromosome number), diverse phenotypic abnormalities, and predisposition to cancer. Little is known about FAM76B and its role in cancer. Other eQTL regulated genes include ZFP57 zinc finger protein (ZFP57), which is a stem cell transcription factor that has recently been found to induce insulin-like growth factor 2 (IGF2) and promote anchorage-independent growth in cancer cells [Tada et al., 2014].
Although this approach of using the secondary phenotype can help to identify genetic variants associated with a primary phenotype, it does not eliminate the potential for false-negative findings when the association with the secondary phenotype is not strong enough to be detected. Specifically, a variant could be associated with lung cancer risk even if it is not associated with a positive family history of lung cancer among first-degree relatives. The difference can be due to various mechanisms that would lead to lung cancer susceptibility. For example, if the effect of the genetic variant is more prominent in the presence of specific environmental exposures, it would not necessarily be associated with family history of lung cancer if the environmental exposures were absent (or present at a minimum level) in past generations. The best example would be the known lung cancer region in Ch15q25, which is thought to be at least partially mediated through smoking behavior, and tends to have stronger association in patients with older age of onset; it is not associated with family history of lung cancer. In this case, this approach would not further inform what was already found in the standard GWAS analysis.
A limitation of our study is that the Replication datasets did not have family history information. In the Replication stage, an exploratory analysis of variants with first-degree family history would potentially lend even greater evidence to a causal relationship between variants and lung cancer. However, our outcome of primary interest was lung cancer in the Replication stage, so this was not considered to be a serious limitation.
Another limitation is the lack of histology information in the largest dataset available from dbGAP. Consequently our stratified analysis by histology did not include this study, potentially compromising the power to detect a statistically significant association for adenocarcinoma, small cell carcinoma, and squamous cell carcinoma.
Prior biological knowledge is a useful tool when investigating the genetics of lung cancer. We have used the well-known association between first-degree family history of lung cancer and lung cancer [Cote et al., 2012] to supplement our investigation of variants related to lung cancer. With the aid of our secondary phenotype analysis, we were able to overcome the strict selection criteria often used in GWAS analysis by giving priority to 537 variants with observed associations with the secondary phenotype. We were then able to identify a subset of 30 variants based on further selection in the Replication stage. This approach, combined with eQTL analysis of identified lung cancer variants in lung tissue, has identified a number of biologically relevant associations that would have been missed by traditional GWAS criteria.
Acknowledgment
This study was funded by the Canadian Cancer Society Research Institute (no. 020214), National Cancer Institute R01 CA149462-02 and Transdisciplinary Research in cancer of the Lung U19 CA148127-01. The scientific development and funding of this project were in part supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON): a NCI Cancer Post-GWAS Initiative. This work was in part supported by the National Institute of Health (USA; grant number CA148127) and earlier sample collection by the Deutsche Krebshilfe (grant number 70-2387). The lung eQTL study at Laval University was supported by the Chaire de pneumologie de la Fondation JD Bégin de l’UniversitéLaval, the Fondation de l’Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the FRQS, the Canadian Institutes of Health Research (MOP–123369), and the Cancer Research Society and Read for the Cure. Y. Bosséis the recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec – Santé(FRQS). Ma’en Obeidat is a Postdoctoral Fellow of the Michael Smith Foundation for Health Research (MSFHR) and the Canadian Institute for Health Research (CIHR) Integrated and Mentored Pulmonary and Cardiovascular Training program (IMPACT).
We thank the following investigators for their contributions to the Central Europe study: Drs. Paolo Boffetta, David Zaridze, Neonilia Szeszenia-Dabrowska, Jolanta Lissowska, Peter Rudnai, Eleonora Fabianova, Dana Mates, Vladimir Bencko, Lenka Foretova and Vladimir Janout. We would also like to thank Li Rita Zhang, Laura Adams, and Zhuo Chen for their assistance of the MSH-PMH study.
References
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne A, Vreede L, Kuiper RP, Bodmer D, Jansen C, Eleveld M, van Erp F, Arkesteijn G, Hoogerbrugge N, van Ravenswaaij C. Mapping of constitutional translocation breakpoints in renal cell cancer patients: identification of KCNIP4 as a candidate gene. Cancer Genet Cytogenet. 2007;179:11–18. doi: 10.1016/j.cancergencyto.2007.07.005. others. [DOI] [PubMed] [Google Scholar]
- Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colditz GA, Manson JE, Hankinson SE. The Nurses’ Health Study: 20-year contribution to the understanding of health among women. J Womens Health. 2007;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- Coté ML, Liu M, Bonassi S, Neri M, Schwartz AG, Christiani DC, Spitz MR, Muscat JE, Rennert G, Aben KK. Increased risk of lung cancer in individuals with a family history of the disease: a pooled analysis from the International Lung Cancer Consortium. Eur. J Cancer. 2012;28:1957–1968. doi: 10.1016/j.ejca.2012.01.038. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby KS, Cuthrell K, Caberto C. Nicotine metabolism in three ethnic/racial groups with different risks of lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:3526–3535. doi: 10.1158/1055-9965.EPI-08-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Hao K, Bossé Y, Nickle DC, Paré PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC. Lung eQTLs to help reveal the molecular underpinnings of asthma. Plost Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BE, Sheppard K, Berndt A, Leme AS, Myers RA, Gignoux CR, Levin AM, Gauderman WJ, Yang JJ, Mathias RA. Integration of mouse and human genome-wide association data identifies KCNIP4 as an asthma gene. PLoS One. 2013;8:e56179. doi: 10.1371/journal.pone.0056179. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. others. [DOI] [PubMed] [Google Scholar]
- Ichimura A, Hirasawa A, Poulain-Godefroy O, Bonnefond A, Hara T, Yengo L, Kimura I, Leloire A, Liu N, Iida K. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature. 2012;483:350–354. doi: 10.1038/nature10798. others. [DOI] [PubMed] [Google Scholar]
- Lamontagne M, Couture C, Postma DS, Timens W, Sin DD, Paré PD, Hogg JC, Nickle D, Laviolette M, Bossé Y. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. Plos One. 2013;8:e70220. doi: 10.1371/journal.pone.0070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Consonni D, Rotunno M, Bergen AW, Goldstein AM, Lubin JH, Goldin L, Alavanja M, Morgan G, Subar AF. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk of adenocarcinoma. AmJ Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Zeng D. Proper analysis of secondary phenotype data in case-control association studies. Genet Epidemiol. 2009;33:256–265. doi: 10.1002/gepi.20377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S, Navarro MGJ, Bossard C, Laurell H, Touriol C, Lacazette E, Prats H. Pivotal role of translokin/CEP57 in the unconventional secretion versus nuclear translocation of FGF2. Traffic. 2009;10:1765–1772. doi: 10.1111/j.1600-0854.2009.00985.x. [DOI] [PubMed] [Google Scholar]
- Momotani K, Khromov AS, Miyake T, Stukenberg T, Somlyo AV. Cep57, a multidomain protein with unique microtubule and centrosomal localization domains. Biochem J. 2008;412:265–273. doi: 10.1042/BJ20071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsees GM, Tamimi RMT, Kraft P. Genome-wide association scans for secondary traits using case-control samples. Genet Epidemiol. 2009;33:717–728. doi: 10.1002/gepi.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeidat M, Miller S, Probert K, Billington CK, Henry AP, Hodge E, Nelson CP, Stewart CE, Swan C, Wein LV. GSTCD and INTS12 regulation and expression in human lung. PLoS One. 2013;8:e74630. doi: 10.1371/journal.pone.0074630. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ. PLINK: a toolset for whole-genome association and population-based linkage analysis. AmJ Hum Genet. 2007;81:559–575. doi: 10.1086/519795. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DB, Rzehak P, Klenk J, Weiland SK. Analysis of case-control data for additional outcomes. Epidemiology. 2007;18:441–445. doi: 10.1097/EDE.0b013e318060d25c. [DOI] [PubMed] [Google Scholar]
- Sauter W, Rosenberger A, Beckmann L, Kropp S, Mittelstrass K, Timofeeva M, Wölke G, Steinwachs A, Scheiner D, Meese E. Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:1127–1135. doi: 10.1158/1055-9965.EPI-07-2840. others. [DOI] [PubMed] [Google Scholar]
- Shi J, Chatterjee N, Rotunno M, Wang Y, Pesatori AC, Consonni D, Li P, Wheeler W, Broderick P, Henrion M. Inherited variation at chromosome 12p13.33 including RAD52 influences squamous cell lung carcinoma risk. Cancer Disc. 2012;2:131–139. doi: 10.1158/2159-8290.CD-11-0246. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape K, Hanks S, Ruark E, Barros-Núñez P, Elliott A, Murray A, Lane AH, Shannon N, Callier P, Chitayat D. Mutations in CEP57 cause Mosaic variegated aneuploidy syndrome. Nat Genet. 2011;43:527–529. doi: 10.1038/ng.822. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Yamaguchi Y, Kinjo T, Song X, Akagi T, Takamura H, Ohta T, Yokota T, Koide H. The stem cell transcription factor ZFP57 induces IGF2 expression to promote anchorage-independent growth in cancer cells. Oncogene. 2014 doi: 10.1038/onc.2013.599. doi:10.1038/onc.2013.599. [DOI] [PubMed] [Google Scholar]
- Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeböller H, Risch A, McKay JD, Wang Y, Dai J. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–4995. doi: 10.1093/hmg/dds334. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thun GA, Imboden M, Ferrarotti I, Kumar A, Obeidat M, Zorzetto M, Haun M, Curjuric I, Couto Alves A, Jackson VE. Causal and synthetic associations of variants in the SERPINA gene cluster with alpha1-antitrypsin serum levels. Plos Genet. 2013;9:e1003585. doi: 10.1371/journal.pgen.1003585. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. AmJ Hum Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- Wain LV, Sayers I, Soler Artigas M, Portelli MA, Zeggini E, Obeidat M, Sin DD, Bossé Y, Nickle D, Brandsma CA. Whole exome re-sequencing implicates CCDC38 and cilia structure and function in resistance to smoking related airflow obstruction. Plos Genet. 2014;10:e1004314. doi: 10.1371/journal.pgen.1004314. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, McKay JD, Rafner T, Wang Z, Timofeeva MN, Broderick P, Zong X, Laplana M, Wei Y, Han Y. Rare variants of large effect in BRCA and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46:736–741. doi: 10.1038/ng.3002. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wang H, Zhao X, Shi Y, Jin M, Wan B, Xu H, Cheng Y, Ge H, Zhang Y. Identification of G-protein-coupled receptor 120 as a tumor-promoting receptor that induces angiogenesis and migration in human colorectal carcinoma. Oncogene. 2013;32:5541–5550. doi: 10.1038/onc.2013.264. [DOI] [PubMed] [Google Scholar]