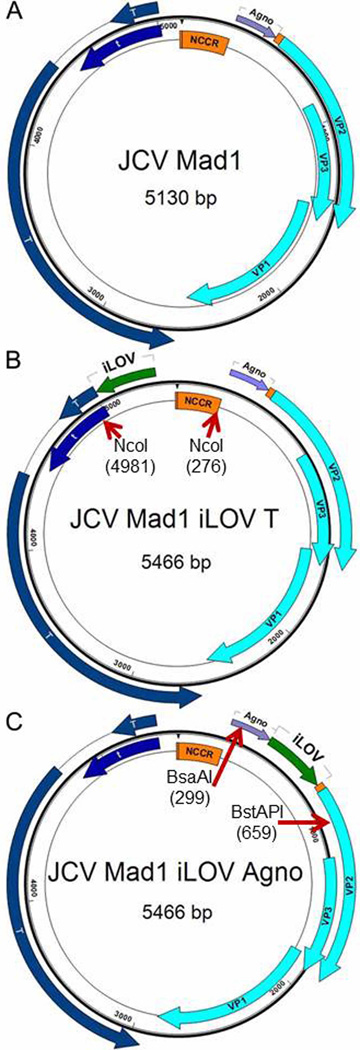
Fig. 1.
Genomic maps of parental JCV strain Mad1 and two improved Light, Oxygen and Voltage sensing domain of plant phototropin (iLOV) constructs: iLOV-Agno and iLOV-T. A: JCV Mad1. JCV Mad1 strain has 5130bp double stranded circular DNA genome which can be divided into non coding control region (NCCR) and coding regions. Coding sequences can be further divided into an early gene group which includes the T and t regulatory protein genes and a late gene group which includes the Agno protein and three viral protein (VP) genes - VP1, 2 and 3. There is a 33bp intercoding region between the end codon of the Agno gene and the start codon of VP2 gene. B: JCV iLOV-T. This construct has a 5466bp genome, as a result of inserting a 336bp improved iLOV gene at position 5013 before the start codon of T gene. The iLOV gene has been optimized for expression in human cell lines. C: JCV iLOV-Agno. This construct has a 5466bp genome, as a result of inserting the 336bp iLOV gene right after the end codon of Agno gene.