Abstract
Purpose
There is a critical need for population-based prospective cohort studies because they follow individuals before the onset of disease, allowing for studies that can identify biomarkers and disease-modifying effects, and thereby contributing to systems epidemiology.
Participants
This paper describes the design and baseline characteristics of an intensively examined subpopulation of the LifeLines cohort in the Netherlands. In this unique subcohort, LifeLines DEEP, we included 1539 participants aged 18 years and older.
Findings to date
We collected additional blood (n=1387), exhaled air (n=1425) and faecal samples (n=1248), and elicited responses to gastrointestinal health questionnaires (n=1176) for analysis of the genome, epigenome, transcriptome, microbiome, metabolome and other biological levels. Here, we provide an overview of the different data layers in LifeLines DEEP and present baseline characteristics of the study population including food intake and quality of life. We also describe how the LifeLines DEEP cohort allows for the detailed investigation of genetic, genomic and metabolic variation for a wide range of phenotypic outcomes. Finally, we examine the determinants of gastrointestinal health, an area of particular interest to us that can be addressed by LifeLines DEEP.
Future plans
We have established a cohort of which multiple data levels allow for the integrative analysis of populations for translation of this information into biomarkers for disease, and which will offer new insights into disease mechanisms and prevention.
Keywords: EPIDEMIOLOGY, PUBLIC HEALTH, GENETICS
Strengths and limitations of this study.
This cohort study is unique in that it collected a wide range of biomaterials (eg, exhaled air and faeces) contemporaneously from fasting individuals.
The LifeLines DEEP cohort is relatively small (n=1539), nevertheless, it will allow for proof-of-concept studies using systems biology approaches.
LifeLines DEEP is an example of a ‘next-generation’ population cohort study—in which multiple molecular data levels are combined with observational research methods.
The data from this study will allow us to construct risk profiles for genetic predisposition to many common diseases and to link these profiles to phenotype information, as well as clinical and immunological parameters.
Extensive questionnaires on, for example, food intake and medical status, will provide data to correct molecular analyses for environmental factors.
Introduction
Many diseases are multifactorial in origin, meaning that they are caused by a combination of genetic and environmental components. To date, a considerable number of genetic variants have been identified that are associated with almost every multifactorial disease or trait.1 These independent genetic factors are often common, occurring frequently in the absence of disease, and therefore cannot yet be used to predict disease. For example, 40 risk loci have been identified for coeliac disease that explain about 54% of disease risk,2 yet there is no clear correlation between carrying these risk alleles and actually developing coeliac disease.3 Thus the question remains: why do some people develop the disease while others are resilient despite carrying many genetic risk alleles? These resilient individuals may provide important clues to disease prevention, but they can only be identified when apparently healthy individuals are followed over time. This highlights the need for prospective cohort studies where life course processes are investigated and determinants of health and disease are identified. An advantage of population-based prospective cohort studies is that they are not specifically targeted to a diseased population and they follow individuals before disease onset, allowing for studies that can identify biomarkers and disease-modifying effects.4 Furthermore, age-related processes that correlate to health and disease can be studied in these cohorts.
LifeLines is a population cohort of over 165 000 participants that covers multiple generations of participating families and focuses on determinants for multifactorial diseases. The cohort includes detailed information on phenotypic and environmental factors, as well as health status.5 6 Moreover, genetic information is available for about 10% of the population. A subset of approximately 1500 LifeLines participants also take part in LifeLines DEEP. These participants are examined more thoroughly, specifically with respect to molecular data, which allow for a more in-depth investigation of the association between genetic and phenotypic variation. For these participants, additional biological materials and information on health status are collected. Subsequently, genome-wide transcriptomics and methylation data are generated, metabolites and biomarkers are measured and the gut microbiome is assessed.
LifeLines DEEP specifically allows for in-depth analysis of gastrointestinal (GI) health-related problems such as irritable bowel syndrome (IBS). This is an important direction for research since GI symptoms are highly prevalent in the general population and have a high impact on quality of life.7 8 IBS is a functional bowel disorder that involves abdominal pain or discomfort and related change in bowel habits.9 Prevalence of IBS in Western countries varies widely among different studies ranging from 4% up to 22%.10 There are, however, no specific tests available to diagnose IBS. The current diagnosis is based on excluding GI diseases and on symptoms using diagnostic criteria such as the Rome III criteria.9
Here we describe the study design and baseline characteristics of the LifeLines DEEP cohort and explain how the collected data can be applied to multiple fields of interest.
Cohort description
LifeLines
Individuals aged 25–50 years were invited by their general practitioner to participate in the LifeLines study. On inclusion, the participants’ family members were also invited to participate in order to obtain information on three generations. At baseline, all participants visited one of the LifeLines Research Sites twice for physical examinations. Prior to these visits, two extensive baseline questionnaires were completed at home. At the first visit, anthropometry, blood pressure, cognitive functioning and pulmonary function as well as other factors were measured (see online supplementary table S1). At the second visit, approximately 2 weeks later, a fasting blood sample was collected. In total, 167 729 participants have been included who will be followed for 30 years.6 Every 18 months, each participant receives a follow-up questionnaire. Additionally, once every 5 years, follow-up measurements of the health parameters are performed.
LifeLines DEEP
From April to August 2013, all participants registered at the LifeLines Research Site in Groningen were invited to participate in LifeLines DEEP, in addition to the regular LifeLines programme. During the participant's second visit to the site, three additional tubes of blood were drawn by one of the LifeLines physicians’ assistants. Exhaled air was also collected during this visit and participants were given instructions for faeces collection at home by one of the LifeLines DEEP assistants. The participants who agreed to collect a faecal sample were also asked to fill in the questionnaire on GI symptoms. Immediately after faecal sample collection, the sample was frozen at −20°C. Faecal samples were collected on dry-ice from the participants’ homes within 2 weeks after the second site visit. On arrival at the research location, the faecal samples were immediately stored at −80°C.
The LifeLines DEEP study was approved by the ethics committee of the University Medical Centre Groningen. All participants signed an informed consent prior to enrolment.
Inclusion
Initially, 1539 participants were included in the LifeLines DEEP study. Of these participants, 78 dropped out: 51 did not complete the second visit to the LifeLines location in time and 27 withdrew from participation. In total, 1461 individuals completed the LifeLines DEEP study. From these participants, we collected additional blood for genetics, methylation and transcriptomics analyses (n=1387); exhaled air for analysis of volatile organic compounds (n=1425); and faecal samples for microbiome and biomarker assessment (n=1248; table 1). Moreover, 1176 GI symptoms questionnaires were returned. For 81% (n=1183) of the participants, we collected all three biomaterials: additional blood, exhaled air and faeces (see online supplementary figure S1). For 11.5% (n=168) of the participants, we collected additional blood and exhaled air, and for 4.4% (n=65) of the participants, we collected exhaled air and faeces. For 3.1% of the participants, we only have additional blood (2.5%, n=36) or exhaled air (0.6%, n=9).
Table 1.
Overview of additional data collected in LifeLines DEEP, including the number of samples, the source biomaterial it originates from and the method of analysis used
| LifeLines DEEP data | n | Source | Methods |
|---|---|---|---|
| Biological ageing | 1387 | Cells from whole blood | FlowFish |
| Biological ageing | 1387 | DNA from whole blood | qPCR and sjTRECs |
| Biomarkers (citrulline, cytokines) | 1387 | Plasma | HPLC, ECLIA |
| Biomarkers (calprotectin, HBD-2, chromogranin A, SCFA) | 1248 | Faeces | ELISA, ELISA, RIA, GC-MS |
| CVD risk score | 1448 | Biochemical measures and questionnaire | Scoring algorithm Framingham Heart Study |
| Functional studies | 1387 | PBMC from whole blood | Various methods |
| Gastrointestinal symptoms | 1176 | Questionnaire | Rome III criteria and Bristol Stool Form Scale |
| Genetics | 1387 | DNA from whole blood | CytoSNP and ImmunoChip, GoNL as imputation reference |
| Metabolomics | 1425 | Exhaled air | GC-tof-MS |
| Metabolomics | 1387 | Plasma | NMR |
| Methylation | 761+ | DNA from whole blood | 450 K chip |
| Microbiome | 1248 | Faeces | 16S rRNA based sequencing |
| Transcriptomics | 1387 | Whole blood (PAXgene) | RNA sequencing |
CVD, cardiovascular disease; ECLIA, electro-chemiluminescence immunoassay; GC-(tof-)MS, gas chromatography-(time of flight-)mass spectrometry; HBD-2, human β defensin 2; HPLC, high-performance liquid chromatography; NMR, nuclear MR; qPCR, quantitative PCR; RIA, radioimmunoassay; SCFA, short chain fatty acids; sjTRECs, signal joint T cell receptor excision circles.
Additional data types
Genome-wide transcriptomics were assessed as a measure of gene expression. We isolated RNA from whole blood collected in a PAXgene tube using PAXgene Blood miRNA Kit (Qiagen, California, USA). The RNA samples were quantified and assessed for integrity before sequencing. Total RNA from whole blood was deprived of globin using GLOBINclear kit (Ambion, Austin, Texas, USA) and subsequently processed for sequencing using Truseq V.2 library preparation kit (Illumina Inc, San Diego, California, USA). Paired-end sequencing of 2×50 bp was performed using Illumina's Hiseq2000, pooling 10 samples per lane. Finally, read sets per sample were generated using CASAVA, retaining only reads passing Illumina's Chastity Filter for further processing. On average, the number of raw reads per individual after QC was 44.3 million. After adapter trimming, the reads were mapped to human genome build 37 using STAR (https://code.google.com/p/rna-star/). Of these, 96% of reads were successfully mapped to the genome. Transcription was quantified on the gene and meta-exon level using BEDTools (https://code.google.com/p/bedtools/) and custom scripts, and on the transcript level using FluxCapacitor (http://sammeth.net/confluence/display/FLUX/Home).
We isolated total DNA from EDTA tubes and profiled genome-wide methylation using the Infinium HumanMethylation450 BeadChip, as previously described.11 In short, 500 ng of genomic DNA was bisulfite modified and used for hybridisation on Infinium HumanMethylation450 BeadChips, according to the Illumina Infinium HD Methylation protocol.
We determined metabolites in exhaled air and blood. Metabolites from exhaled air were measured by a combination of gas chromatography and time-of-flight mass spectrometry (GC-tof-MS), as described previously.12 13 In short, the exhaled air sample was introduced in a GC that separates the different compounds in the mixture. Subsequently, the compounds were introduced into the MS to detect and also to identify the separated volatile organic compounds. The metabolites in plasma were measured using the nuclear MR (NMR) method, as described by Kettunen et al.14
Genotyping of genomic DNA was performed using both the HumanCytoSNP-12 BeadChip15 and the ImmunoChip, a customised Illumina Infinium array.16 Genotyping was successful for 1385 samples (CytoSNP) and 1374 samples (IChip), respectively. First, SNP quality control was applied independently for both platforms. SNPs were filtered on MAF above 0.001, a HWE p value >1e−4 and call rate of 0.98 using Plink.17 The genotypes from both platforms were merged into one data set. For genotypes present on both platforms, the genotypes were put on missing in the case of non-concordant calls. After merging, SNPs were filtered again on MAF 0.05 and call rate of 0.98, resulting in a total of 379 885 genotyped SNPs. Next, these data were imputed based on the Genome of the Netherlands (GoNL) reference panel.18–20 The merged genotypes were prephased using SHAPEIT221 and aligned to the GoNL reference panel using Genotype Harmonizer22 in order to resolve strand issues. The imputation was performed using IMPUTE223 V.2.3.0 against the GoNL reference panel. We used a MOLGENIS compute24 imputation pipeline to generate our scripts and monitor the imputation. Imputation yielded 8 606 371 variants with Info score ≥0.8. In addition, HLA type was established via the Broad SNP2HLA imputation pipeline.25
We collected several types of cells, including lymphocytes and granulocytes, for assessment of telomere length as a measure for ageing. We are now optimising the FlowFish method of telomere measuring as described by Baerlocher et al.26 In addition, peripheral blood mononuclear cells (PBMCs) were collected and stored at −80°C for future functional studies.
Faecal samples were collected in order to study the gut microbiome. Gut microbial composition was assessed by 16S rRNA gene sequencing of the V4 variable region on the Illumina MiSeq platform according to the manufacturer's specifications.27 Reads were quality filtered and taxonomy was inferred using a closed reference Operational Taxonomic Unit-picking protocol against a preclustered GreenGenes database, as implemented by QIIME (V.1.7.0 and V.1.8.0).28 29 Moreover, faecal aliquots were stored for future analysis of GI-health-related biomarkers.
In addition, phenotypic data were collected on GI health symptoms by means of the Rome III criteria questionnaire9 and the Bristol Stool Form Scale.30
We collected and stored plasma for future analysis of disease and ageing-related biomarkers such as circulating microRNAs.
Analyses of baseline characteristics, quality of life, GI symptoms and qualitative food intake
For each participant, a risk score for cardiovascular disease (CVD) was calculated according to the scoring algorithm developed by the Framingham Heart Study.31 The CVD risk score ranges from ≤−3 to ≥18 and is calculated based on gender, age, high-density lipoprotein, total cholesterol, systolic blood pressure, smoking status and presence or absence of diabetes.
We calculated quality of life scores based on the RAND 36-item Short Form Health Survey scoring version I by calculating eight summary scores, and the mental and physical component score.32 33 The summary scores range from zero to 100 points, where 100 represents the best quality of life. The mental and physical component scores were transformed to have a mean of 50 and a SD of 10 compared to the reference population, as described by Ware et al.33 34
Occurrence of functional bowel disorders was assessed via the Rome III criteria.9 Participants with self-reported Crohn's disease, ulcerative colitis and caeliac disease were excluded from this analysis.
Data on habitual dietary intake were collected via a validated food frequency questionnaire developed by the division of Human Nutrition of Wageningen University.35
Mean and SDs for the baseline characteristics and quality of life scores were calculated. Statistical programmes R (V.3.0.1) and IBM SPSS Statistics (V.20) were used for analyses and for constructing the figures.
Baseline characteristics of study participants
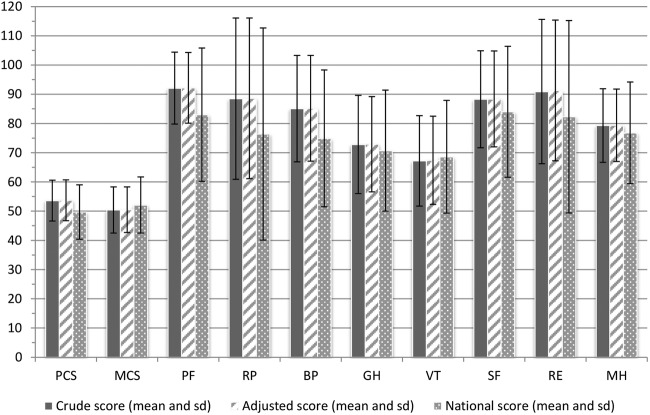
Over a period of 6 months, 1539 participants enrolled in the LifeLines DEEP study. Slightly more women (n=903, 58.7%) than men (n=636, 41.3%) were included (table 2). The age of the participants ranged from 18 to 86 years, with a mean age of 44. Mean BMI was 25.2 kg/m2. On average, total cholesterol level and blood glucose level both were 5 mmol/L. Average blood pressure was lower in women (116/68 mm Hg) compared to men (124/74 mm Hg). Among women, the percentage of current smokers was slightly lower (18.3%) than in men (19.5%). The Framingham risk score for cardiovascular disease was, on average, 5.7 for women and 8.6 for men, corresponding to a 3% and 7% risk of a first cardiovascular event, respectively (table 2).31 In our cohort, the quality of life score was lowest for vitality (mean(SD): 67.2(15.5)) and highest for physical functioning (mean(SD): 92.1(12.3)) (figure 1 and online supplementary table S2). The data on age, BMI and blood level parameters were normally distributed, whereas the data on CVD risk score and QoL components deviated from normality.
Table 2.
Baseline characteristics of LifeLines DEEP by gender, including smoking, age, BMI, cholesterol level, glucose level, blood pressure and Framingham risk score for cardiovascular disease
| Characteristic |
Men | Women | ||
|---|---|---|---|---|
| n | 636 | 903 | ||
| Smoking status | Current | % | 19.5 | 18.3 |
| Former | % | 29.9 | 28.5 | |
| Never | % | 47.0 | 48.0 | |
| Age | mean (SD) | 44.0 (13.9) | 43.3 (13.8) | |
| BMI | mean (SD) | 25.4 (3.5) | 25.0 (4.7) | |
| Total cholesterol (mmol/L) | mean (SD) | 5.0 (1.0) | 5.0 (1.0) | |
| HDL cholesterol (mmol/L) | mean (SD) | 1.3 (0.3) | 1.7 (0.4) | |
| Glucose level (mmol/L) | mean (SD) | 5.1 (0.7) | 4.9 (0.7) | |
| Systolic blood pressure (mm Hg) | mean (SD) | 123.5 (12.4) | 115.6 (13.4) | |
| Diastolic blood pressure (mm Hg) | mean (SD) | 73.6 (9.6) | 68.4 (8.3) | |
| CVD risk score | mean (SD) | 8.6 (8.9) | 5.7 (7.0) | |
BMI, body mass index; CVD, cardiovascular disease; HDL, high-density lipoprotein.
Figure 1.
Mean and SD of crude and adjusted quality of life scores, 2 component scores and 8 group scores in the LifeLines DEEP population (n=1539) compared to a national sample of the Dutch population.34 42 Adjusted score is adjusted for gender and age. PCS, physical component score; MCS, mental component score; PF, physical functioning; RP, role-physical; BP, bodily pain; GH, general health; VT, vitality; SF, social functioning; RE, role-emotional; MH, mental health.
Analysis of 1176 GI symptoms questionnaires identified 409 participants with functional bowel disorders (figure 2). Prevalence of IBS in our cohort was 21% (n=249). Another 13% (n=160) of participants fulfilled criteria for functional bloating (9%, n=108), functional constipation (3%, n=37) or functional diarrhoea (1%, n=15). Two-thirds of the participants (n=767) did not meet the Rome III criteria for functional bowel disorders. Moreover, 4% (n=51) of the participants answered that they never experience any GI symptoms (figure 2).
Figure 2.
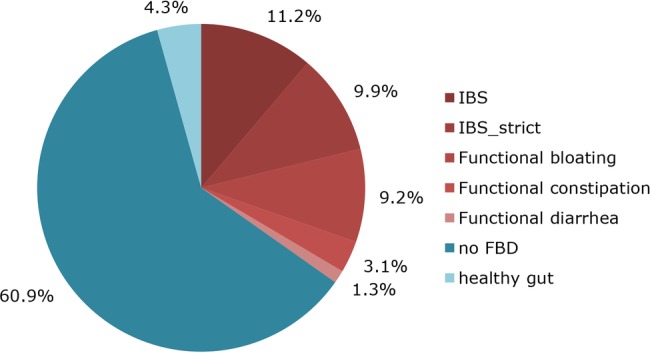
Functional bowel disorders in the LifeLines DEEP cohort based on Rome III criteria (n=1176). IBS (Irritable Bowel Syndrome): pain or discomfort at least 2–3 days/month, IBS_strict: pain or discomfort more than 1 day/week, FBD, functional bowel disorder, healthy gut, lowest possible score on Rome III questionnaire.
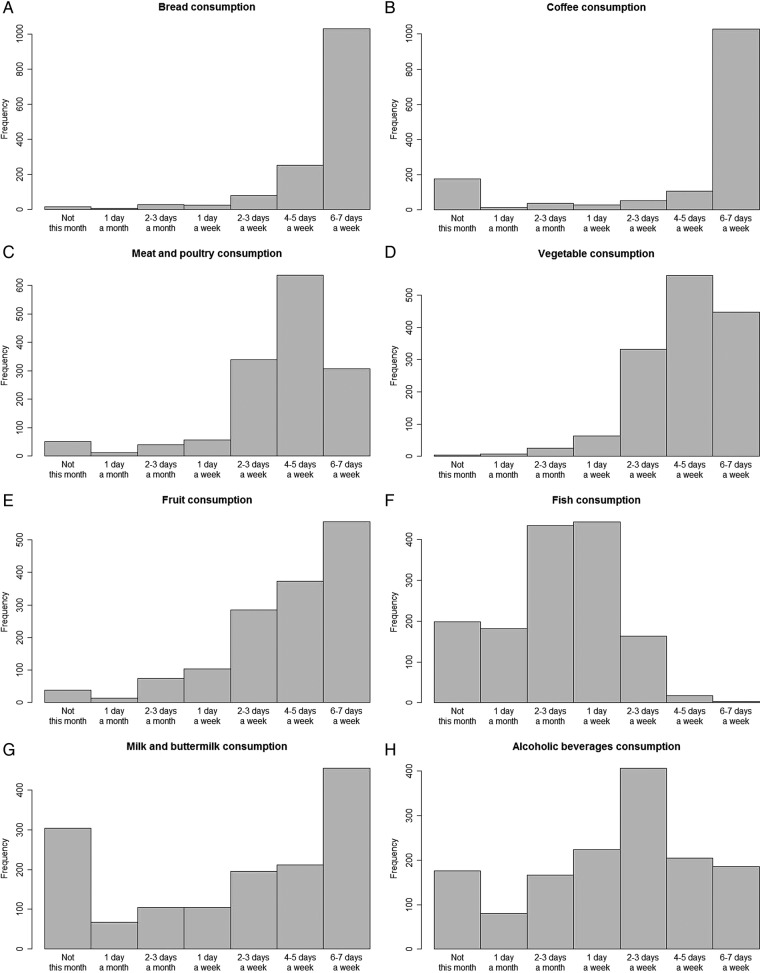
Analysis of the frequency of intake of major food groups showed a subdivision into three main categories. The first category contained food groups that were consumed daily, bread and coffee, for example (figures 3A,B). The second category contained food groups for which consumption ranged from daily to a few days per week. Examples of these food groups include meat, vegetables and fruit (figures 3C–E). The third category included food groups that were consumed on a weekly to monthly basis only, fish, for example (figure 3F). For other food groups, such as milk and alcoholic beverages, the intake varied greatly (figures 3G, H). These frequency data will later be combined with portion sizes and the Dutch food composition table (NEVO 2006, RIVM, Bilthoven) to estimate nutrient intake in grams per day.
Figure 3.
Qualitative intake of (A) bread, (B) coffee, (C) meat and poultry, (D) vegetables, (E) fruit, (F) fish, (G) milk and buttermilk and (H) alcoholic beverages, in LifeLines DEEP (n=1539). Bars represent: ‘not this month’, ‘1 day/month’, ‘2–3 days/month’, ‘1 day/week’, ‘2–3 days/week’, ‘4–5 days/week’ and ‘6–7 days/week’.
For all individuals, additional biomaterials were collected (see online supplementary figure S1) for future system epidemiological studies36 integrating multilevel ‘omics’ data with environmental, physical and epidemiological data to provide a deeper and more detailed view of the LifeLines DEEP population. These biomaterials include plasma to examine the concentration of metabolites, peripheral blood mononuclear cells to determine genome-wide transcription and methylation profiles, exhaled air to analyse volatile organic compounds and faeces to establish composition of the gut microbiome. Moreover, genetic data has been generated for all individuals, allowing for the construction of genetic risk profiles for a wide variety of common diseases. These multiple data levels will provide rich opportunities for future research into the molecular underpinnings of human health and disease, as well as for research into the interaction between molecular and environmental components including behaviour, sociodemographic factors and analysis of specific subgroups. For example, the analysis of microbiota composition in relation to ageing revealed associations to several taxa, and highlights the importance of correcting for age in microbiome studies (figure 4).
Figure 4.
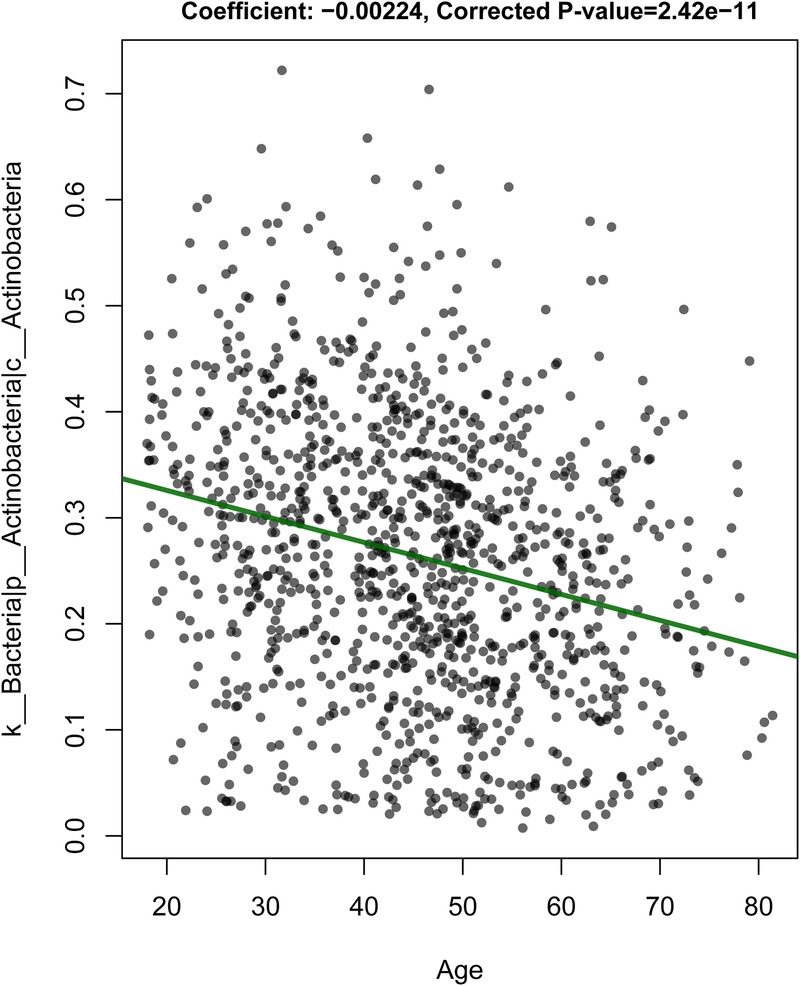
Change in abundance of Actinobacteria on ageing.
Findings to date
One area of particular interest is the domain related to GI health. We therefore studied the prevalence of IBS in LifeLines DEEP. We identified IBS in 21% of participants. These data should be interpreted with caution as our diagnosis is based solely on participant's responses to a Rome III criteria questionnaire, and results may therefore be slightly inflated. Nevertheless, our result is consistent with previous suggestions that almost 25% of the population encounters irritable bowel symptoms over the course of their lifetimes.37 Its prevalence in our cohort confirms that IBS is a common disease and thus research aimed at improved diagnosis and treatment will benefit society. GI symptoms are multifactorial, making large cohorts necessary to study them in more detail. For IBS, in particular, there is an urgent need to develop biomarkers that are predictive of disease. We have selected six GI-health-related biomarkers and developed a multidomain biomarker panel that can distinguish patients with IBS from healthy controls and that correlates well to GI symptom severity in patients with IBS (Mujagic Z 2015, submitted for publication). The biomarker panel was developed in the Maastricht IBS cohort (which currently includes 400 cases and 200 healthy controls, recruitment is ongoing) and validated in the LifeLines DEEP cohort. In addition, we studied the volatile organic compound (VOC) profile from exhaled air in both cohorts and compared IBS cases with controls (Baranska A 2015, submitted for publication). We identified a novel breath biomarker of 16 VOCs that distinguishes patients with IBS from healthy controls and correlates significantly with the presence of GI symptoms. Furthermore, we are collaborating with a large Genome-Wide Association Study on the identification of the genetic architecture of IBS.38 Several chromosomal regions of suggestive significance were identified in individual cohorts. Integrative meta-analysis of this data is currently ongoing. Moreover, we are working on the analysis of food intake in patients with IBS versus healthy controls.
Despite the high prevalence of IBS, the quality of life in our study population in general was higher than in a random selection of the Dutch population as reported by Aaronson et al.39 This might be due to age and gender differences, since the national sample included 56% men with a mean(SD) age of 47.6(18) years,39 compared to 41% men with a mean(SD) age of 44.6(13.8) years in the LifeLines DEEP cohort. Secular changes may also play a role, since the national survey was conducted more than 15 years ago.
LifeLines DEEP is also a unique, independent data source. For approximately 1500 individuals, we will be able to construct genetic risk profiles for predisposition to many common diseases based on genome-wide association data. Next, we will be able to link these risk profiles to phenotype information, as well as clinical, immunological and other parameters. Using this information, we may already be able to compare high-risk individuals with and without disease symptoms to generate hypotheses on resilient individuals. Recently, Ricaño-Ponce et al studied the genetics of 14 immune-mediated diseases and identified single nucleotide polymorphisms (SNPs) specifically affecting the expression of long non-coding RNAs in these diseases (Ricaño-Ponce I 2015, submitted for publication). At the same time, LifeLines DEEP also allows for integration across different data levels to study, for example, the association between molecular and phenotypic data to increase our understanding of pathogenic mechanisms.40 For instance, on analysing data from the LifeLines DEEP cohort, we found associations of bacterial taxonomies to age (figure 4), and to BMI and blood lipid levels (Fu J 2015, in press at Circulation Research). Furthermore, Smolinska et al performed extensive analysis on confounding factors such as smoking and BMI on VOCs analysis (Smolinska A 2015, manuscript in preparation).
Strengths and limitations
In the design of a population cohort study, it is important to balance breadth (the number of samples included) and depth (the amount of phenotypic data). LifeLines is a large prospective cohort that includes more than 165 000 individuals and measures several thousand phenotypes ranging from biochemical parameters, physical measurements, psychosocial characteristics and environmental factors, to detailed information on health status. However, the cohort was not set up to include molecular data levels for the study of health and disease in human populations. With LifeLines DEEP, we are performing a pilot study of additional deep molecular measurements in 1500 individuals, using biomaterials from different domains that were all collected contemporaneously from fasting individuals. Although the LifeLines DEEP cohort is relatively small, it will allow for proof-of-concept studies into systems epidemiology. LifeLines DEEP is unique in that it has exhaled air measurements from all individuals and a level of information that, to our knowledge, is rarely present in other population-based cohorts. Additionally, both the collection of cells for telomere length measurements and further functional studies, and the faecal sample collection, are unique. LifeLines DEEP will not only contribute to a better understanding of the association between genetic variation and molecular function, but can also be integrated with other population cohorts that have similar molecular data. In particular, the collection and analysis of faecal material is crucial given increasing evidence that the gut microbiome can play an important role in health and disease.41 Nevertheless, harmonisation and linking of data across multiple cohorts might be needed to achieve critical numbers. The Biobanking and Biomolecular Research Infrastructure in the Netherlands (BBMRI-NL)42 and Europe43 will allow for such studies. LifeLines DEEP has been designed to study exposures in detail whereas data on disease heterogeneity is limited. Combining our exposure-driven-data-collection cohort with disease-specific and tissue-specific-data-collection cohorts, such as the Netherlands Cohort Study on Cancer, could offer even more insight into disease mechanisms.44 45
Collaboration
We have established a cohort of which multiple data layers allow for integrative analysis of populations for translation of this information into biomarkers for disease and which will provide new insights into disease mechanisms and prevention. We encourage collaborations with researchers from other cohort studies to work on the above aspects with increased sample size. The data from the LifeLines DEEP cohort will be available via LifeLines.6 Researchers can apply for data and biomaterial by submitting a proposal to the LifeLines Research Office (LLscience@umcg.nl). Detailed information on the measured variables can be found in the online LifeLines data catalogue (http://www.lifelines.net). All proposals will be reviewed on scientific quality and methodology by the LifeLines scientific board.
Acknowledgments
The authors would like to thank the LifeLines participants and the staff of the LifeLines study site, Groningen, for their collaboration. The authors would also like to thank the LifeLines DEEP research assistants, Wilma Westerhuis-van der Tuuk, Marc Jan Bonder, Astrid Maatman, Mathieu Platteel, Kim de Lange and Debbie van Dussen, for their practical and analytical work. The authors thank Jackie Senior and Kate McIntyre for editing our manuscript. Furthermore, the authors would like to thank The Target project (http://www.rug.nl/target) for providing the computer infrastructure and the BigGrid/eBioGrid project (http://www.ebiogrid.nl) for sponsoring the imputation pipeline implementation.
Footnotes
Contributors: CW, LF, JAMD, RPS and AZ were involved in the conception and design of the study. EFT, JAMD, GH, AB, ZM, MAS, PD, MCC and SS contributed to development of methods and data collection. EFT, AZ, AMM and EJMF were involved in data analysis and interpretation. EFT and SZ drafted the work. All the authors have critically revised this article and approved the final version to be published.
Funding: This project was funded by a Top Institute Food and Nutrition Wageningen grant GH001 to CW, the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL) grant RP3 to LF and an ERC advanced grant ERC-671274 to CW. SZ holds a Rosalind Franklin fellowship (University of Groningen). MCC has a postdoctoral fellowship from the Spanish Fundación Alfonso Martín Escudero.
Competing interests: None declared.
Ethics approval: The LifeLines DEEP study was approved by the ethics committee of the University Medical Centre Groningen, document number METC UMCG LLDEEP: M12.113965. All participants signed an informed consent form prior to study enrolment.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Researchers can apply for data and biomaterial by submitting a proposal to the LifeLines Research Office (LLscience@umcg.nl). The LifeLines website provides information on the application process (http://www.lifelines.net).
References
- 1.Welter D, Macarthur J, Morales J et al. . The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001–6. 10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar V, Wijmenga C, Withoff S. From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Semin Immunopathol 2012;34:567–80. 10.1007/s00281-012-0312-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romanos J, Rosén A, Kumar V et al. . Improving coeliac disease risk prediction by testing non-HLA variants additional to HLA variants. Gut 2014;63:415–22. 10.1136/gutjnl-2012-304110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, Bailey-Wilson JE, Collins FS. Genes, environment and the value of prospective cohort studies. Nat Rev 2006;7:812–20. 10.1038/nrg1919 [DOI] [PubMed] [Google Scholar]
- 5.Stolk RP, Rosmalen JG, Postma DS et al. . Universal risk factors for multifactorial diseases: LifeLines: a three-generation population-based study. Eur J Epidemiol 2008;23:67–74. 10.1007/s10654-007-9204-4 [DOI] [PubMed] [Google Scholar]
- 6.Scholtens S, Smidt N, Swertz MA et al. . Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int J Epidemiol 2014. 10.1093/ije/dyu229 [DOI] [PubMed] [Google Scholar]
- 7.El-Serag HB, Olden K, Bjorkman D. Health-related quality of life among persons with irritable bowel syndrome: a systematic review. Aliment Pharmacol Ther 2002;16:1171–85. 10.1046/j.1365-2036.2002.01290.x [DOI] [PubMed] [Google Scholar]
- 8.Gralnek IM, Hays RD, Kilbourne A et al. . The impact of irritable bowel syndrome on health-related quality of life. Gastroenterology 2000;119:654–60. 10.1053/gast.2000.16484 [DOI] [PubMed] [Google Scholar]
- 9.Longstreth GF, Thompson WG, Chey WD et al. . Functional bowel disorders. Gastroenterology 2006;130:1480–91. 10.1053/j.gastro.2005.11.061 [DOI] [PubMed] [Google Scholar]
- 10.Drossman DA, Camilleri M, Mayer EA et al. . AGA technical review on irritable bowel syndrome. Gastroenterology 2002;123:2108–31. 10.1053/gast.2002.37095 [DOI] [PubMed] [Google Scholar]
- 11.Harris RA, Nagy-Szakal D, Pedersen N et al. . Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis 2012;18:2334–41. 10.1002/ibd.22956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boots AW, van Berkel JJ, Dallinga JW et al. . The versatile use of exhaled volatile organic compounds in human health and disease. J Breath Res 2012;6:27108 10.1088/1752-7155/6/2/027108 [DOI] [PubMed] [Google Scholar]
- 13.Baranska A, Tigchelaar E, Smolinska A et al. . Profile of volatile organic compounds in exhaled breath changes as a result of gluten-free diet. J Breath Res 2013;7:037104 10.1088/1752-7155/7/3/037104 [DOI] [PubMed] [Google Scholar]
- 14.Kettunen J, Tukiainen T, Sarin AP et al. . Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet 2012;44:269–76. 10.1038/ng.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolmans GH, Werker PM, Hennies HC et al. . Wnt signaling and Dupuytren's disease. N Engl J Med 2011;365:307–17. 10.1056/NEJMoa1101029 [DOI] [PubMed] [Google Scholar]
- 16.Trynka G, Hunt KA, Bockett NA et al. . Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet 2011;43:1193–201. 10.1038/ng.998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boomsma DI, Wijmenga C, Slagboom EP et al. . The Genome of the Netherlands: design, and project goals. Eur J Hum Genet 2014;22:221–7. 10.1038/ejhg.2013.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deelen P, Menelaou A, van Leeuwen EM et al. . Improved imputation quality of low-frequency and rare variants in European samples using the ‘Genome of The Netherlands’. Eur J Hum Genet 2014;22:1321–6. 10.1038/ejhg.2014.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francioli LC, Menelaou A, Pulit SL et al. . Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014;46:818–25. 10.1038/ng.3021 [DOI] [PubMed] [Google Scholar]
- 21.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 2013;10:5–6. 10.1038/nmeth.2307 [DOI] [PubMed] [Google Scholar]
- 22.Deelen P, Bonder MJ, van der Velde KJ et al. . Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res Notes 2014;7:901 10.1186/1756-0500-7-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–70. 10.1534/g3.111.001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byelas H, Dijkstra M, Neerincx P et al. . Scaling bio-analyses from computational clusters to grids. IWSG; 2013. http://ceur—ws.org/Vol—993/paper2.pdf [Google Scholar]
- 25.Jia X, Han B, Onengut-Gumuscu S et al. . Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 2013;8:e64683 10.1371/journal.pone.0064683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baerlocher GM, Vulto I, de Jong G et al. . Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc 2006;1:2365–76. 10.1038/nprot.2006.263 [DOI] [PubMed] [Google Scholar]
- 27.Gevers D, Kugathasan S, Denson LA et al. . The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15:382–92. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, Kuczynski J, Stombaugh J et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeSantis TZ, Hugenholtz P, Larsen N et al. . Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–72. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Donnell LJ, Virjee J, Heaton KW. Detection of pseudodiarrhoea by simple clinical assessment of intestinal transit rate. BMJ 1990;300:439–40. 10.1136/bmj.300.6722.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Agostino RB Sr, Vasan RS, Pencina MJ et al. . General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 2008;117:743–53. 10.1161/CIRCULATIONAHA.107.699579 [DOI] [PubMed] [Google Scholar]
- 32.Ware JE Jr, Kosinski M et al. . Comparison of methods for the scoring and statistical analysis of SF-36 health profile and summary measures: summary of results from the Medical Outcomes Study. Med Care 1995;33:AS264–79. [PubMed] [Google Scholar]
- 33.Ware JE, Kosinski M, Keller S. SF-36 physical and mental summary scales: a user's manual. Boston: New England Medical Center, 1994. [Google Scholar]
- 34.Ware JE, Gandek B, Kosinski M et al. . The equivalence of SF-36 summary health scores estimated using standard and country-specific algorithms in 10 countries: results from the IQOLA Project. International Quality of Life Assessment. J Clin Epidemiol 1998;51:1167–70. 10.1016/S0895-4356(98)00108-5 [DOI] [PubMed] [Google Scholar]
- 35.Siebelink E, Geelen A, de Vries JH. Self-reported energy intake by FFQ compared with actual energy intake to maintain body weight in 516 adults. Br J Nutr 2011;106:274–81. 10.1017/S0007114511000067 [DOI] [PubMed] [Google Scholar]
- 36.Haring R, Wallaschofski H. Diving through the ‘-omics’: the case for deep phenotyping and systems epidemiology. OMICS 2012;16:231–4. 10.1089/omi.2011.0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones R, Lydeard S. Irritable bowel syndrome in the general population. BMJ 1992;304:87–90. 10.1136/bmj.304.6819.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ek WE, Reznichenko A, Ripke S et al. . Exploring the genetics of irritable bowel syndrome: a GWA study in the general population and replication in multinational case-control cohorts. Gut 2014. Published Online First. [DOI] [PubMed] [Google Scholar]
- 39.Aaronson NK, Muller M, Cohen PD et al. . Translation, validation, and norming of the Dutch language version of the SF-36 Health Survey in community and chronic disease populations. J Clin Epidemiol 1998;51:1055–68. 10.1016/S0895-4356(98)00097-3 [DOI] [PubMed] [Google Scholar]
- 40.Dumas ME, Kinross J, Nicholson JK. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology 2014;146: 46–62. 10.1053/j.gastro.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 41.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012; 489:242–9. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 42.Brandsma M, Baas F, Bakker PIW et al. . How to kickstart a national biobanking infrastructure—experiences and prospects of BBMRI-NL. Nor Epidemiol 2012;21:143–8. [Google Scholar]
- 43.Wichmann HE, Kuhn KA, Waldenberger M et al. . Comprehensive catalog of European biobanks. Nat Biotechnol 2011;29: 795–7. 10.1038/nbt.1958 [DOI] [PubMed] [Google Scholar]
- 44.Bishehsari F, Mahdavinia M, Vacca M et al. . Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol 2014;20:6055–72. 10.3748/wjg.v20.i20.6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogino S, Lochhead P, Chan AT et al. . Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol 2013;26:465–84. 10.1038/modpathol.2012.214 [DOI] [PMC free article] [PubMed] [Google Scholar]