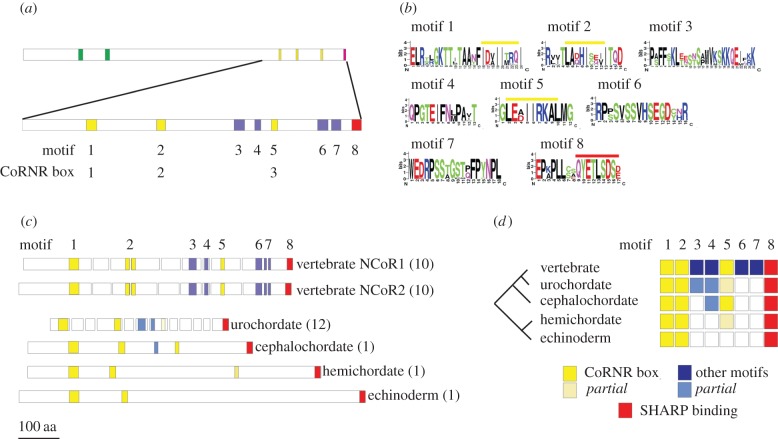
Figure 1.
The NCoR-family conserved motifs and exon structure. (a) The NCoR-family proteins in the vertebrates typically contain two SANT domains (green bar), followed by three CoRNR boxes, nuclear receptor interaction motifs (yellow bars) and a carboxy-terminal SHARP-binding motif (red bar). Alignment of vertebrate NCoR1 and NCoR2 sequences identifies a further four conserved motifs (blue bars, lower diagram). Full sequence alignments are in the electronic supplementary material, figure S1. (b) Identity of conserved vertebrate sequences using the motif notation. Yellow bars overlie the consensus CoRNR box motif L/I.x.x.I/H.I.x.x.x.I/L [50,51] that is embedded in each of motifs 1, 2 and 5. The C-terminal SHARP-binding sequence is overlined in red as part of motif 8. (c) Exon organization of the 3′ end of representative NCoR-family genes. The regions encoding the motifs have been mapped onto the relevant exons maintaining the colour scheme in (a). In parentheses is the number of exons in this region of the gene. The C-terminal motifs are encoded by one large exon encoding 843 amino acids in the sea urchin, but 12 exons encoding 365 amino acids in the sea squirt. (d) Summary of C-terminal motif acquisition across the representative deuterostome panel. All contain motifs 1,2 (CoRNR boxes 1 and 2) and 8, but motif 5 (the third CoRNR box) is not present in the echinoderm and incomplete in the hemichordate and urochordate. Two of the four vertebrate specific motifs (3 and 4) are represented by partial motifs in the urochordate and cephalochordate (motif 4 only). Sequence alignments of the motifs are in the electronic supplementary martial, figure S2.