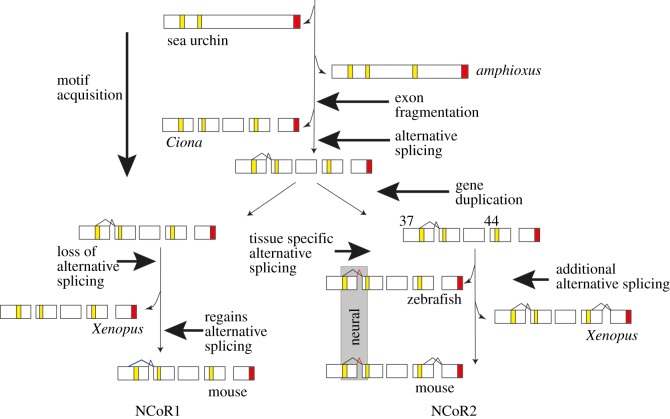
Figure 6.
The pathway to diversity for the NCoR family of co-repressors. Strongylocentrotus purpuratus (sea urchin) encodes two CoRNR boxes, but this increases to three in the cephalochordate amphioxus. Further motifs, identified by similarity to those in vertebrates, are found in the urochordate Ciona intestinalis. An additional CoRNR box motif will increase the range of nuclear receptors to which the co-repressor can bind. While the C-terminal interaction domains are encoded by a single exon in amphioxus they are encoded by at least 10 exons in Ciona and the vertebrates (for clarity, not all exons are shown). The ability to restore exon 37b alternative splicing in NCoR1 suggests that alternative splicing of this exon arose in the NCoR-family gene before duplication, which happened during the vertebrate genome duplication event after the divergence of Ciona and the vertebrates [69]. Of the two resulting paralogues, NCoR1 lost the alternative splicing of exon 37b by point mutation of the splice donor. Mammals, however, have recovered the capacity for alternative splicing to generate an isoform that lacks the first CoRNR box but which employs a different splice donor (blue lines). The alternative splicing of NCoR2 exon 37 is apparent in the teleost zebrafish, where, like mammals such as the mouse, the presence of the long form (red lines) including CoRNR box1 is tissue-specific, being found only in neural tissue. In addition Xenopus and the mouse undergo alternative splicing to generate isoforms of exon 44 [27]. Zebrafish lack the capacity for exon 44 alternative splicing. CoRNR boxes are in yellow and the SHARP domain in red.