Abstract
Discovered in 1909 by Retzius and described mainly by morphology, the cytoplasmic droplet of sperm (renamed here the Hermes body) is conserved among all mammalian species but largely undefined at the molecular level. Tandem mass spectrometry of the isolated Hermes body from rat epididymal sperm characterized 1511 proteins, 43 of which were localized to the structure in situ by light microscopy and two by quantitative electron microscopy localization. Glucose transporter 3 (GLUT-3) glycolytic enzymes, selected membrane traffic and cytoskeletal proteins were highly abundant and concentrated in the Hermes body. By electron microscope gold antibody labelling, the Golgi trafficking protein TMED7/p27 localized to unstacked flattened cisternae of the Hermes body, as did GLUT-3, the most abundant protein. Its biogenesis was deduced through the mapping of protein expression for all 43 proteins during male germ cell differentiation in the testis. It is at the terminal step 19 of spermiogenesis that the 43 characteristic proteins accumulated in the nascent Hermes body.
Keywords: membrane traffic, cryosection electron microscopy localization, subcellular fractionation, quantitative proteomics, light microscope localization
1. Introduction
Discovered in 1909 [1], the cytoplasmic droplet of epididymal sperm is seen as a small bulge of cytoplasm (2 µm in diameter) positioned at the neck or connecting piece of the flagellum [2–4]. The presence and migration of the cytoplasmic droplet along the sperm flagellum from the neck to the annulus [2,5,6] occur in proximal regions of the epididymal duct where motility is initiated [7,8].
By electron microscopy (EM) of the droplet, flattened unstacked cisternae are abundant, which, along with dilated and elongated membranous profiles and vesicles of different sizes, concentrate and segregate to one pole of the spherical structure [9]. The flattened cisternae of the cytoplasmic droplet are derived from the fragmentation and unstacking of the Golgi apparatus of late spermatids, as first deduced by morphology and Golgi protein localization [9], and later confirmed by Moreno et al. [10].
The cytoplasmic droplet is needed to initiate motility as deduced from a comparison of sperm with or without droplets [5]. By mass spectrometry of five bands excised from SDS PAGE of isolated cytoplasmic droplets, 105 largely soluble proteins (including glycolytic enzymes) were characterized that were proposed to regulate the initiation of motility through the generation of ATP [11]. However, the major morphological structures in the droplet, including the characteristic flattened internal membranes [9], were not retained in the isolated structure and integral membrane proteins were largely missing.
The biogenesis of the cytoplasmic droplet occurs in the testis at the last step of germ cell differentiation, step 19 of spermiogenesis [9]. Testis-expressed proteins affect droplet formation. For example, spermatid maturation 1 (Spem1) expressed in late steps of germ cell differentiation in the testis is needed for normal droplet formation and fertility [12]. The 15Lox gene that expresses organelle degradation enzyme, 15-lipoxygenase in male germ cells of the testis, is required for epididymal sperm maturation, droplet migration and morphology, normal fertility, and normal litter size [13]. With respect to the Golgi origin of the internal flattened cisternae of the structure, a recent detailed analysis of Golgi apparatus in differentiating germ cells of the testis revealed a subset of Golgi proteins in the forming structure at step 19 [14]. However, the contribution of proteins from other membrane sources and other cellular structures was not assessed. This was the objective of this study.
Using a procedure that retains the internal membranes of the isolated cytoplasmic droplet and a methodology for quantitative protein characterization of all proteins including integral membrane proteins by tandem mass spectrometry, 1511 abundant proteins have been characterized in this study. Through antibody-based localizations of 58 proteins, their expression during germ cell differentiation in the testis could be compared with localization to the cytoplasmic droplet of sperm in the epididymis, including the 1318 proteins characterized previously for male germ cell Golgi apparatus [14].
Based on these data, the hypothesis that the cytoplasmic droplet originates in the late spermatid of the testis to coordinate membrane trafficking with the initiation of sperm motility in the epididymis is supported. We propose that the cytoplasmic droplet be renamed the Hermes body. Hermes is a winged god of transitions and boundaries, and with a physical attribute to male virility. The Hermes body is deduced to regulate, through its makeup of enzymes and internal membranes and cytoskeletal constituents, the transition of an immotile, unfertile to motile, fertile sperm in the epididymis.
2. Material and methods
2.1. Animals
All animals used in this study were maintained on a 12 L/12 D cycle in the animal facility and fed ad libitum. The procedures for animal use were done in accordance with the guidelines of the McGill Animal Care Committee.
2.2. Hermes body isolation and characterization
Adult Sprague Dawley rats (350–450 g) were purchased from Charles River Laboratories Canada (St Constant, Quebec, Canada). For each isolation procedure, 10 rats were used. After anaesthesia, the testes were removed from the scrotum following intracardiac saline perfusion. The epididymis was removed from the testis by cutting through the rete testis. The initial segment, caput and corpus down to its mid area were removed and placed in ice-cold buffer (50 mM Tris–HCl pH 7.4, 25 mM KCl, containing either 1 mM PMSF, 200 K units of aprotinin per millilitre of buffer and 1 µM leupeptin per millilitre of buffer or 0.5 mM Pefabloc; Sigma). The epididymides were removed from the buffer, placed on a paraffin block on ice and diced into 1 mm pieces. Diced epididymides were placed in a Falcon tube with 15–20 ml of ice-cold buffer and then vortexed at medium to high speed 5 × 10 s, being returned to the ice for a few seconds between each vortex. The suspension was poured through a metal sieve and then through a 150 µm Nitex net to remove connective tissue. A milky suspension of sperm was collected in a 30 ml Corex tube and centrifuged at 1500g for 15 min (Avanti rotor R-20, 3500 r.p.m. (1500g). The pellet (P1) was put aside on ice. The supernatant (S1) was centrifuged at 150 000g for 15 min at 4°C (Ti60 rotor, 36 000 r.p.m., 100 000g). The supernatant (S2) was discarded. The pellet (P2) was combined with P1, and re-suspended in 5 ml of ice-cold buffer with a glass rod. The suspension was passed through a 20 gauge needle five times, and then centrifuged at 150g for 10 min at 4°C to give a pellet (P3) and supernatant (S3). S3 (4.5 ml) was placed above a sucrose step gradient made of 2 ml each of 0.6 M, 0.8 M, 1.0 M and 1.2 M sucrose (in ice-cold buffer), with the refractometer readings 19.2, 25.0, 31.0 and 35.5, respectively. This was centrifuged for 90 min at 40 000 r.p.m. in an SW-40 rotor (202 000g) with the brake on. The interface between 0.8 and 1 M sucrose (fraction 3) was collected with a needle and syringe and designated the cytoplasmic droplet/Hermes body fraction.
Fraction 3 was enriched in the Golgi marker enzyme UDP-galactose ovomucoid galactosyl transferase 61.3-fold ± 22.5 (mean ± s.d., n = 4) over the sperm homogenate.
2.3. Routine electron microscopy processing of in situ epididymis and isolated subcellular fractions
For routine EM analysis, four adult male Sprague Dawley rats were anaesthetized with sodium pentobarbital and their epididymides fixed by perfusion through the abdominal aorta with 2.5% glutaraldehyde in 0.2 M sodium cacodylate buffer containing 0.1% calcium chloride, pH 7.4 [2]. After fixation (10 min), the tissue was removed, trimmed and left in fixative for 2 h. It was then postfixed in potassium ferrocyanide reduced osmium tetroxide for 1 h, after which it was dehydrated in alcohol, acetone and embedded in Epon. Sections were cut with a diamond knife and stained with uranyl actetate and lead citrate, and examined with a Philips 400 EM, Tecnai 12 120 kV TEM and Tecnai G2 F20 Cryo-STEM (FEI Inc.).
For isolated Hermes bodies, portions of samples from n = 3 isolated fractions were processed for EM analysis. Two millilitre of isolated subcellular fractions (100–150 µg protein) in ice-cold buffer were mixed with 2 ml of fixative (5% glutaraldehyde in 0.2 M sodium cacodylate buffer containing 0.1% calcium chloride, pH 7.4) on ice in a chemical fume hood. The Hermes body fractions were collected onto filter membranes (Millipore nitrocellulose filter 0.45 mm HA), washed with 0.1 M cacodylate buffer (pH 7.4 containing 5% sucrose) three to four times, incubated with tannic acid (1% tannic acid Mallinckrodt CN No 1764 in 0.1 M cacodylate buffer pH 7.4) for 1 h, washed with cacodylate buffer containing 1% sodium sulfate, placed in 100 mM maleate buffer pH 5.7, incubated with uranyl acetate (6% uranyl acetate in 0.1 M sodium maleate, pH 5.7) for 2 h on ice in the fume hood, washed with maleate buffer, dehydrated through a series of graded alcohols (70%, 80%, 90%, 95% and 100% ethanol) for 10 min for each step, and then dissolved in 100% acetone for 1–2 h. The samples were embedded in Epon resin and the blocks were then cut and sectioned (90–100 nm), stained with uranyl acetate and lead citrate and examined with a Tecnai 12 120 kV TEM equipped with a Gatan 792 Bioscan CCD Camera (Gatan Inc., Pleasanton, CA, USA).
2.4. Electron microscope tomography
Samples were prepared as described above. However, thicker sections (approx. 250 nm) were cut for electron tomography and transferred onto carbon-coated copper grids. Images were collected on a Titan Krios microscope operated at 300 kV using a Gatan Ultrascan 4kx4 k CCD camera. For electron tomography, data collection was done at an electron dose of approximately 1500 electrons/Å2 per tomogram. Focusing was done on an adjacent area in order to minimize electron dose exposure. In total, over 30 tomograms were collected at different magnifications ranging between 20 and 50 k. Tilt series were taken using the FEI software in the angular range between −64° and +64° with 2o increments. For the estimated sample thickness, this would be sufficient for a resolution of 2 nm, following the Crowther formula. Reconstruction of three-dimensional volumes was done using the IMOD software suite [15]. The final tomograms were binned three times in order to increase the signal-to-noise ratio. Three-dimensional rendering was done using Chimera [16].
2.5. Tandem mass spectrometry
The methodologies described by Gilchrist et al. [17] and Bell et al. [18] were followed. The raw data were processed in pipeline format [19] to generate a peaklist of all tandem MS by employing Distiller followed by Mascot Cluster. The Mascot search results were then parsed into the in-house relational database termed CellMapBase [20], scored for protein identifications and grouped to present a minimum set of protein identifications [21] to account for all tandem MS assigned at 95% confidence. The concatenated peaklist was searched against a copy of the National Center for Biotechnology Information non-redundant database (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nr.gz) (release: NCBI nrdb 2008). For generation of the minimum list of proteins [21], protein identifications were grouped based on redundant peptide assignments taking into account redundancies that arise due to homologous sequences, truncated or partial sequences, alternatively spliced proteins, strain-specific allelic variation or redundant assignments of tandem MS. In this process, peptides were assigned as unique to an identification or shared between two or more identifications. For quantification, redundant peptide counting (spectral counts) was performed essentially as described previously [17,18,22,23]. In the present case, peptides were grouped to their cognate proteins based on the gel resolved sample and shared peptides were apportioned to the cognate proteins based on the proportion of unique peptides. For comparison among different subcellular fractions isolated, the percentage of total peptides for each of n = 3 or n = 4 biological repeats were averaged to represent the percentage total peptide counts for each subcellular fraction (electronic supplementary material, table S1).
2.6. Data analysis
As indicated above, all protein identifications made by peptides assigned by Mascot at the 95% confidence level (false positive rate approx. 1.5% estimated by searching a randomized copy of the database for the legacy data [17]) were tabulated into a relational database (CellMapBase) for further manipulations. The list of identified proteins is shown in the electronic supplementary material, table S1. The non-redundant list (electronic supplementary material, table S1) of proteins identified in this study was finalized as a consequence of curation (see below), first by literature search via the NCBI Gi number to categorize all identifications into 1 of 22 functional categories as described [17] and then to assign to each a representative protein name for equivalent proteins as determined by sequence identity [24]. Functional categories: biosynthetic cargo, blood + other cells, calcium transport/binding, coat, cytoskeleton, detoxification, GTPase, likely contaminants, lysosome, metabolism, mitochondria, nucleus, peroxisome, plasma membrane, proteasome/ubiquitin, protein modification, protein synthesis/folding, signalling, tethering/docking/fusion, traffic, trypsin and unknown.
2.7. Hierarchical clustering
Hierarchical clustering was done using the Cluster 3.0 program (http://bonsai.ims.u-tokyo ac.jp/∼mdehoon/software/cluster/) [25], and clusters were viewed by the Java Treeview program (http://jtreeview.sourceforge.net/) [26]. Excel was used for all data analysis.
2.8. Light microscope immunohistochemistry
The testes and epididymides of adult rats used for light microscope (LM)-immunohistochemistry (IHC) and in some cases LM-immunofluorescence (IF) were fixed by perfusion through the abdominal aorta for 10 min, after which the tissues were left overnight in fresh fixative [2] or by simple immersion in Bouin's for 24 h (n = 4 for each) at room temperature. Other rats were fixed by perfusion or immersion (n = 2 for each) with Zinc Fixative [27] (BD Biosciences, Cat 550523; Mississauga, Ontario, Canada).
Upon removal from the animal, the testes and epididymides were sectioned along their long axis to form two roughly equal halves. After fixation, all tissues were subsequently placed in 70% alcohol for several days before being dehydrated and embedded in paraffin.
Paraffin sections were cut at a 5 µm thickness and mounted on ‘Posi-Plus’ slides (Fisher Scientific Company; Ottawa, Ontario, Canada). Sections of both tissues were deparaffinized with Histoclear (Fisher brand 22-143975; Fisher Scientific) and rehydrated in a series of 100, 95, 80, 70 and 50% ethanol solutions, 0.3 M glycine, and phosphate-buffered saline (PBS).
Following rehydration, immunostaining was performed on Bouin-fixed tissue with the Envision+ System-HRP (DAB, diaminobenzidine) anti-rabbit Kit (Cat K4010; Dako Canada, Mississauga, Ontario, Canada) and a wash buffer solution containing 0.05 M Tris, 0.3 M NaCl and 0.1% Tween 20, pH 7.4. Dilutions for each primary antibody (using 5% bovine serum albumin (BSA) in PBS) were optimized and fell within a range of 1 : 100–1 : 500, and were incubated on the slides at room temperature for 1.5 h. Slides were washed 10 × 1 min and incubated with the secondary antibody (Envision+ kit), for 60 min at room temperature. The slides were again washed for 10 × 1 min, and incubated with the DAB solution from the Envision+ kit (time of incubation was optimized for each protein, and fell into the range of 5–30 s). The reaction product was visualized as a brownish colour with the intensity varying according to a given antibody. The sections were counterstained for 10 s in methylene blue, washed and quickly dehydrated through graded ethanol solutions to Histoclear. Coverslips were mounted onto the slides with Permount.
Negative controls for all experiments consisted of substituting PBS for primary antibody. Negative control experiments were performed using the above protocol, but without primary antibody. Non-immune sera (a preimmune antiserum to enthoprotin) were also substituted for primary antibody as control with no immunoreactivity detected. When available (e.g. anti-calnexin), preimmune sera were also tested and shown to have no detectable immunoreactivity.
2.9. Immunofluorescence
Sections were deparaffinized in hexane (Fisher Scientific), rehydrated in a graded ethanol series and washed in distilled water followed by 50 mM Tris-buffered saline (TBS), pH 7.4. Sections were incubated for 3 h at room temperature with primary antibody diluted as above in TBS. Sections were washed with TBST (TBS + 0.1% Tween 20), blocked for 20 min in a 2% casein solution and incubated for 30 min at room temperature with Alexafluor 594-labelled goat anti-rabbit IgG antibody (Invitrogen Canada) diluted 1 : 500 in TBST. Samples were washed with TBST, rinsed in TBS and counterstained for 1–3 min at room temperature with 300 nM 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen Canada) in TBS. Samples were rinsed in TBS, and coverslips were mounted using Prolong Gold antifade reagent (Invitrogen Canada). Sections were examined and photographed on a Zeiss Axioskop 2 motorized LM equipped with variable intensity FluorArc epifluoresence mercury lighting and AxioCam HR colour digital camera (Carl Zeiss Canada; Montreal, Quebec, Canada). Controls were done as indicated above.
For LM-IF of tissue sections, the results are based on 10 slides with an additional three as controls. Four antibodies were examined by LM-IF.
2.10. Electron microscopy immunogold labelling of epididymis
Two adult male Sprague Dawley rats were anaesthetized with sodium pentobarbital and their epididymides fixed by perfusion through the abdominal aorta with 0.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M PBS (pH 7.4). After removal of the epididymis, the initial segment region was trimmed into small pieces (0.5 mm3), immersed for 2 h in the above fixative at 4°C, washed three times in 0.15 M PBS (pH 7.4), and then treated with PBS. Cryo-ultrathin sections of the small pieces of tissue were obtained following a modified procedure of Tokuyasu [28] and Zierold [29]. The tissue pieces were transferred onto a metal stub, rapidly frozen by plunging into liquid nitrogen, and stored in liquid nitrogen before cryo-ultramicrotomy. The stub was mounted on the specimen holder of an Ultracut-E ultramicrotome (Reichert) equipped with an FCS cryosystem. Cryo-ultrathin sections (70–100 nm thick) were cut from the frozen pieces in a cryo chamber at −70°C with a dry glass knife. The sections were picked up with a gold loop filled with a 2.3 M sucrose solution, transferred onto formvar-coated 200 mesh copper TEM grids, and kept on ice in a drop of PBS and 2.0% BSA/2% casein/0.5% ovalbumin (BCO).
Ultrathin sections of the initial segment of the epididymis were mounted on 300 mesh, formvar-coated nickel grids (Canemco, Montreal, Quebec, Canada). Each grid was floated for 5 min on a drop of 2% glycine in PBS, followed by 10 min on a drop of 2% bovine casein-ovo albumin (BCO), and then incubated for 1 h on 15 µl drops of primary rabbit antibodies to glucose transporter 3 (GLUT-3) and TMED7/p27 in 1 : 1, 1 : 5 and 1 : 10 dilutions with BCO. Sections were washed six times for 5 min each in PBS followed by a 2% BCO block for 5 min. The grids were then incubated for 20 min with a secondary goat anti-rabbit IgG antibody conjugated to 12 nm colloidal-gold (Jackson ImmunoResearch 1 : 20 dilution in BCO). The sections were subjected to six 5 min washes in PBS, followed by six 5 min washes in distilled water. Sections were counterstained with uranyl oxalic acetate stain (pH 7) in water for 5 min, followed by three 1 min washes with distilled water. Sections were protected with a 12 min incubation in a layer of 2% methylcellulose (Anachemia, Montreal, Quebec, Canada).
2.11. Quantification of gold decorated antibody labelling of cryosections of epididymal Hermes bodies
Electron microscopy of immunolabelled cryo sections was conducted using a Tecnai 12 120 kV TEM and Tecnai G2 F20 Cryo-STEM (FEI Inc.). Sections were randomly sampled with the only criteria being presence of gold particles in cross, longitudinal or oblique sections of the Hermes body. Micrographs were taken at magnification ranging from 30 000× to 60 000× with an AMT XR80C CCD Camera System on the Tecnai 12 and a Gatan Ultrascan 4000 4 k × 4 k CCD Camera System Model 895 on the Tecnai F20. Grids prepared with no primary antibody but incubated in gold-conjugated secondary antibody were used as controls.
A total of 10 sections were examined at different dilutions from each of two different animals and at five different dilutions. A total of 290 electron micrographs were analysed, of which 30 were controls.
Gold particle quantification was conducted as follows, with gold particles counted as individual particles when seen individually or when several were closely clustered together. To reveal gold particle distribution over the plasma membrane, longitudinal sections of the sperm flagellum were used. Gold particles in the area of the plasma membrane were scored over several demarcated 12 nm increments placed on either side of the plasma membrane (intra and extracellular). A 72 nm span (36 in each direction) on either side of the plasma membrane was determined as being the area where the most gold particles were positioned. This was determined for GLUT-3 from cross-sectional micrographs of the sperm flagellum where only plasma membrane surrounding an enclosed flagellum were found. To define a reference ‘membrane area’ 12 nm gold particles along a cross section of the flagellum were counted both intracellularly and extracellularly. Gold particles were counted individually, with each particle of clusters being counted as one. A 72 nm area with the flagellum's membrane at its centre was determined to be the area of maximum gold distribution along the cross section of the flagellum. This span of 72 nm was then used as a reference area for gold quantification in micrographs of Hermes bodies allowing classification of gold particles as being located in five distinct areas: cross and oblique sections of plasma membrane, intracytoplasmic (cisternae), and cross and oblique sections of flagellar components. The total number of gold particles was then counted in order to calculate percentage of gold distribution in each category on the various electron micrographs. Sections with no primary antibody were used as controls.
2.12. In situ sperm immunofluorescence
Epididymides were dissected from adult rats (n = 2), and sperm were removed from the caput and corpus regions. Immediately sperm were placed in fixative containing 2% paraformaldehyde and 0.2% glutaraldehyde in PBS for 45 min at room temperature. Samples were centrifuged at low speed (500g for 4 min at room temperature), and the supernatant was removed. Samples were washed (3 × 2 min in PBS), and the auto-fluorescence was quenched by incubating the samples in 50 mM ammonium chloride for 10 min at room temperature. The sperm were washed (3× 2 min in PBS), and then blocked and permeabilized for 45 min at room temperature in PBS containing 0.2% Fish Skin Gelatin (Sigma) and 0.05% Triton X-100. Samples were washed again, and incubated with antibodies to GLUT-3 (diluted 1 : 500 in PBS containing 0.2% fish skin gelatin) for 1 h at room temperature. After washing, the sperm were then incubated with goat anti-rabbit Alexa fluor 546 (diluted 1 : 1000) and phalloidin fluor 488 nm (Molecular Probes; diluted 1 : 200) for 45 min at room temperature. Samples were again washed, and finally incubated with DAPI for 2 min at room temperature. Following a final wash, the sperm were placed on a microscope slide, and coverslips were mounted using Prolong Anti-fade Gold mounting media (Invitrogen). Images were acquired using a LSM 700 series microscopy system (Carl Zeiss) fitted with a plan-apochromat 63×/1.40 oil immersion objective in sequential scanning mode with the pinhole set to obtain an optical section of about 0.8 µm in all channels (approx. 1 Airy unit). DAPI was excited with a 405 nm laser diode, and the emitted fluorescence was filtered through a 420–480 nm band-pass filter. For phalloidin, a 488 nm Argon ion laser was used, and emitted fluorescence was filtered through a 505–530 nm band-pass. Alexa fluor 546 was excited with a 561 nm DPSS laser; the emitted fluorescence was filtered through a 585–690 nm band-pass.
3. Results
3.1. The isolated Hermes body retains internal flattened cisternae encompassed by a plasma membrane
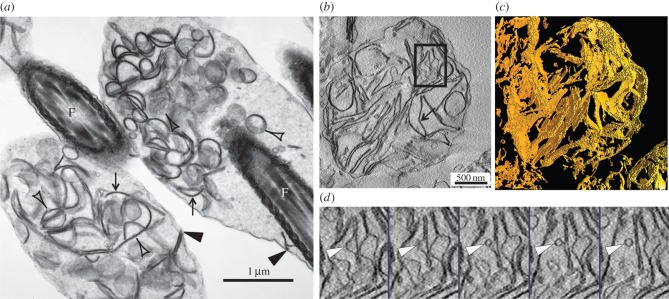
Electron microscopy revealed an identical morphology of the Hermes body of epididymal sperm in situ (figure 1a) to that of its isolated counterpart (figure 1b). A plasma membrane surrounded both, and each contained, as the near exclusive organelle, numerous flattened, straight and curved cisternal elements segregated to one pole, but not forming a stacked configuration. Dilated membranous profiles of different shapes and sizes, as well as numerous 50–60 nm spherical vesicles, were also evident (figure 1a,b). Tomographical three-dimensional analyses of the isolated Hermes body revealed that the cisternae were flattened and sheet-like in appearance (figure 1c). Small vesicles (50–60 nm) were evident (figure 1d; electronic supplementary material, SMov.1). Furthermore, close contiguities were often observed between the edges of adjacent cisternae and the plasma membrane (figure 1a,b). Hence subsequent interrogation of the protein make-up of the Hermes body isolated here should provide insight into the protein make-up of these internal membranes.
Figure 1.
Correspondence of in situ and isolated Hermes body by EM tomography. (a) EM of in situ and (b) isolated Hermes body of epididymal sperm of (IS). Flattened cisternae (arrows), vesicular budding (clear arrowheads). Edges of cisternae in proximity to plasma membrane (dense arrowheads). (c) Tomography of isolated Hermes body reveals sheet-like unstacked flattened cisternae. (d) Series of successive slices (approx. 2.8 nm) of the enclosed area of figure 1b revealing a vesicle adjacent to a cisterna. F, flagellum.
3.2. Cell map strategy to characterize Hermes body proteins
The Hermes bodies from spermatozoa obtained from proximal regions of adult rat epididymides were characterized by tandem mass spectrometry (electronic supplementary material, table S1). The tandem mass spectra from these samples were combined with those from germ cell-enriched Golgi fractions from adult rat testis homogenates [14] and the resource of tandem mass spectra from endoplasmic reticulum (ER), Golgi and Golgi-derived COPI vesicle proteins from adult rat liver [17] (electronic supplementary material, figure S1). Full-length protein sequences were deduced and assigned to each sample based on the proportion of tandem mass spectra for the protein in each sample. The total number of proteins in the resource is 2922, of which 1151 are found in the Hermes body. For functional relevance, all proteins were allocated into 22 categories (electronic supplementary material, figure S2, derived from table S1) designed for interpreting proteomic data of isolated subcellular fractions of the secretory pathway [17]. As well, electronic supplementary material, table S2 indicates all proteins of the Hermes body in order of their abundance.
Hierarchical clustering of the data of electronic supplementary material, table S1 readily revealed abundant proteins that were enriched or unique to the isolated Hermes bodies, or in common with those in testis Golgi fractions and diminished in liver ER and Golgi fractions (electronic supplementary material, figure S3A). Subsequent selection of proteins for detailed LM-IHC and -IF localizations are indicated in electronic supplementary material, table S3 with the protein distributions in the isolated fractions shown in electronic supplementary material, figure S3B, and the proteins localized by IHC, IF or immuno-EM indicated in electronic supplementary material, figure S3C in comparison with those proteins whose localization was already studied in [14].
3.3. Concentration and localization of TMED/p24 membrane trafficking proteins in flattened cisternae of Hermes bodies supports a Golgi identity
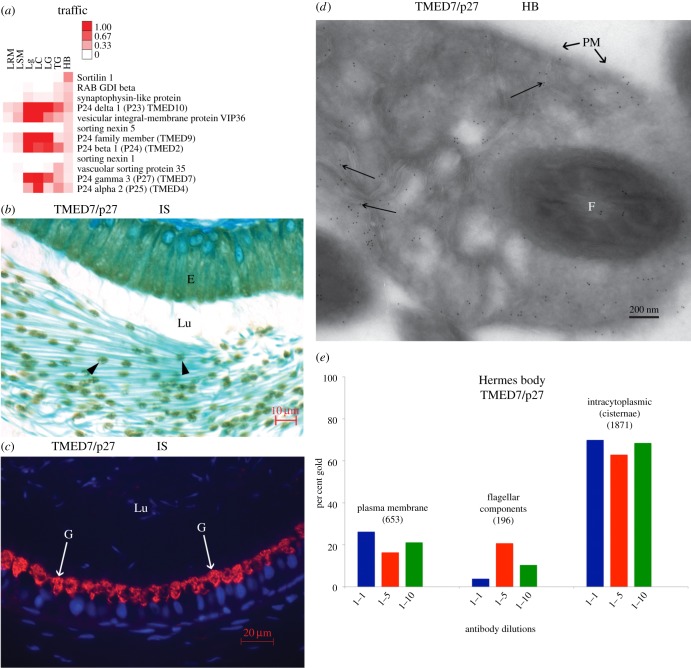
Several TMED (p24 family) proteins for membrane traffic (figure 2a) were concentrated in isolated Hermes bodies. TMED7/p27 is Golgi restricted and expressed at all steps of male germ cell differentiation in the testis [14]. In epididymis, the protein shows a Hermes body segregation in sperm (figure 2b) and Golgi localization in epithelial epididymal cells (figure 2c). By EM immunogold labelling, TMED7/p27 was concentrated in flattened cisternae of the Hermes body (figure 2d) of epididymal sperm in situ. Quantification indicated 67 ± 4% gold particles (mean ± s.d., n = 3) over the flattened cisternae, 21 ± 5% on the plasma membrane of the Hermes body and 12 ± 9% considered as non-specific over flagellar components (figure 2e; electronic supplementary material, table S4). Negligible gold particles were seen without primary antibody. Hence, TMED7/p27 is characterized as a Golgi protein localized to the internal membranes of the Hermes body of epididymal sperm representing the first documentation of this protein to the structure and confirming a Golgi identity [9].
Figure 2.
Hermes bodies localization of TMED7/p27 by LM and EM in situ. (a) Heat map of the 12 most abundant proteins of the Traffic category. (b) IHC shows the localization of TMED7/p27 in epididymal Hermes bodies. (c) IF reveals Golgi reactivity of epididymal epithelial cells for TMED7/p27. However, germ cell immunoreactivity is not seen due to the low sensitivity of the IF protocol. (d) EM immunolocalization of TMED7/p27. Gold particles (arrows) over flattened cisternae, plasma membrane and flagella of sperm. (e) Gold particle quantification in cryosections of Hermes body of sperm. The total number of gold particles scored over the plasma membrane, intracytoplasmic (cisternae) or flagella are indicated above each histogram representing different dilutions of anti-TMED7/p27. E, epithelium; F, flagellum; G, Golgi reactivity; HB, Hermes body; IS, initial segment; Lu, lumen; PM, plasma membrane.
3.4. Protein abundance assigns a major function in glucose uptake and glycolysis
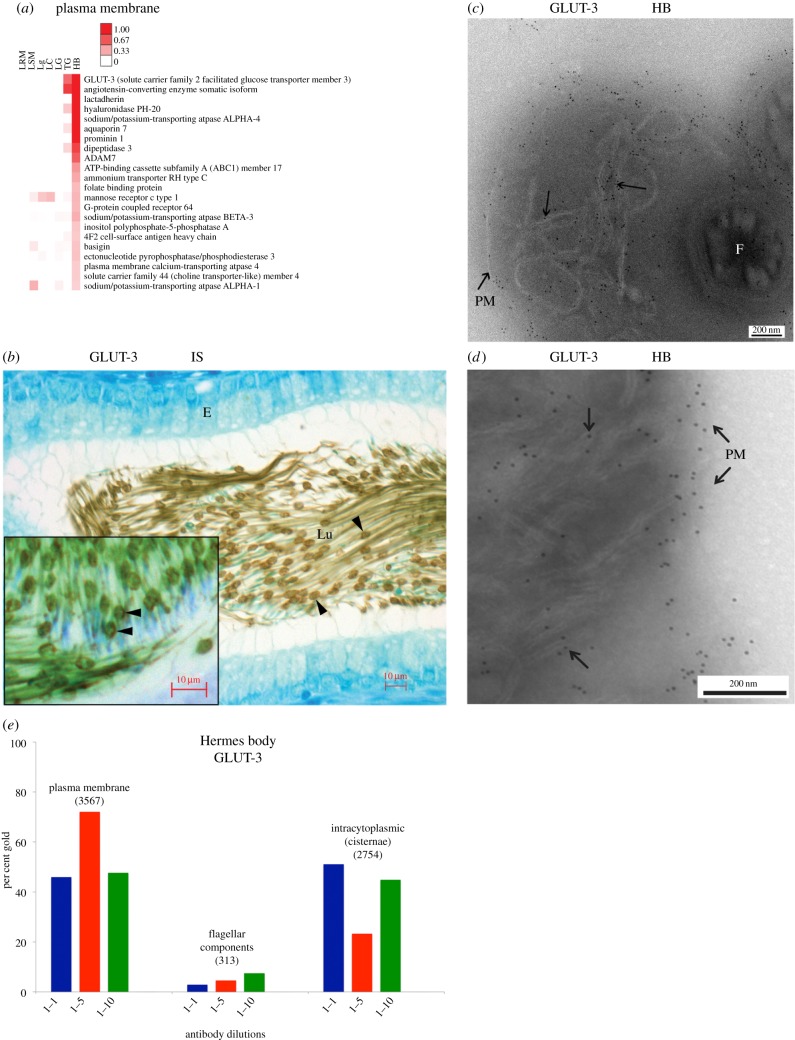
The most abundant protein of the isolated Hermes body (electronic supplementary material, table S2) was the high-affinity GLUT-3. Categorized to plasma membranes (figure 3a), GLUT-3 is a known sperm flagellar plasma membrane protein, identical to neuronal GLUT-3 [30]. Other known sperm plasma membrane proteins detected in the isolated Hermes body included aquaporin 7 [27], angiotensin-converting enzyme [31,32], PH20 [33], dipeptidase 3 [34], mannose receptor [35], ABC1 member 17 [36], folate binding protein [37] and the highly abundant sodium potassium ATPase subunit alpha 4, a germ cell-derived known sperm plasma membrane protein, essential for motility and fertility [38,39]. ADAM 7 was prominent, as were 11 other family members (electronic supplementary material, table S2) all known to be sperm associated [40–42]. Several of these proteins have been characterized in proteomic studies of sperm [43–45]. These were not seen by proteomics of isolated droplets by Yuan et al. [11] due to their use of two-dimensional gels that cannot resolve membrane proteins.
Figure 3.
In situ localization of GLUT-3. (a) Heat map of 22 proteins in plasma membrane category (ordered by abundance and averaged for each subcellular fraction). (b) LM-IHC of GLUT-3 in Hermes body of epididymal sperm and (inset) reaction restricted to one pole. (c,d) EM cryosections of Hermes body of sperm in IS immunolabelled with anti-GLUT-3. Gold particles associated with PM and flattened cisternae. (e) Gold particle labelling at primary antibody dilutions indicated was scored over the plasma membrane, intracytoplasmic (cisternae) or flagellar components as shown in the electronic supplementary material, table S4. The total number of gold particles scored is indicated above each histogram. Abbreviations as in figure 2.
LM-IHC for GLUT-3 revealed localizations in small focal elliptical dilations defined as the Hermes bodies (figure 3b), with immunoreactivity concentrated to one pole (figure 3b, inset). Although less concentrated, immunoreactivity was also observed along the entire midpiece of the sperm flagellum (figure 3b). Immunogold labelling of GLUT-3 on ultrathin cryosections of in situ initial segment epididymal tissue revealed a major concentration of gold particles over the plasma membrane of the Hermes body; gold label is also concentrated over the internal flattened cisternae (figure 3c,d). As shown in figure 3e and electronic supplementary material, table S4, 55 ± 15% (mean ± s.d., n = 3) of all gold particles are on the plasma membrane, with intracellular cisternal GLUT-3 labelling at 40 ± 15% and non-specific gold labelling to flagellar components at 5 ± 2%. Negligible gold particles are found in sections without primary antibody (electronic supplementary material, table S4). The localization of a subset of GLUT-3 to the same internal membranes as TMED7/p27 is consistent with a membrane trafficking function for the internal membranes.
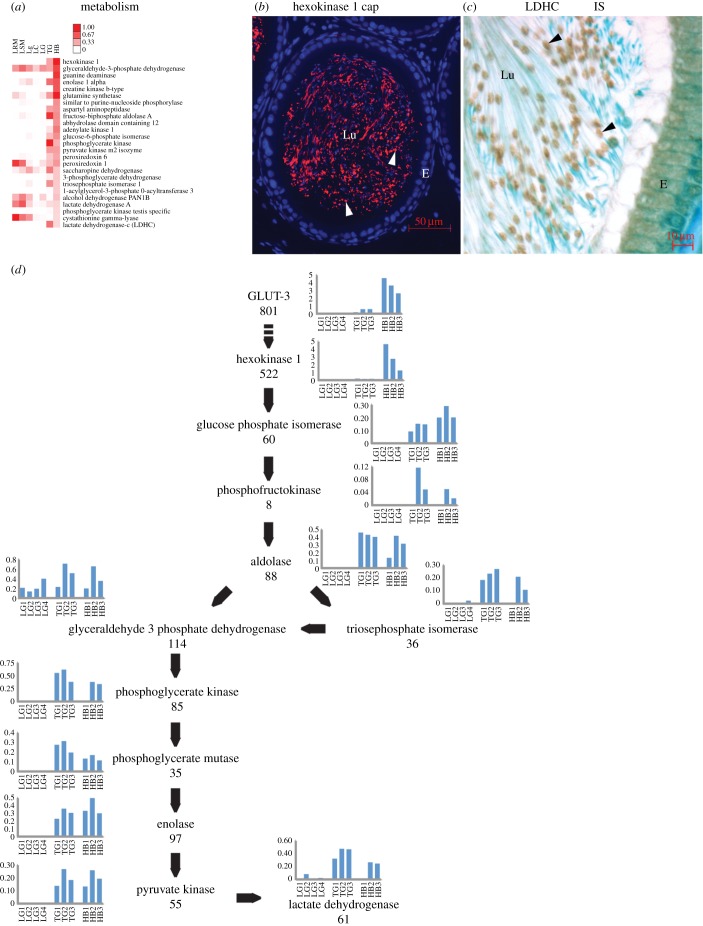
The second most abundant protein of the isolated Hermes body is the first enzyme in glycolysis, Hexokinase 1 (electronic supplementary material, table S2). This is the most abundant protein in the metabolism category (figure 4a). Hexokinase 1 localizes to the Hermes body of sperm in the epididymal lumen (figure 4b). Lactate dehydrogenase C the 25th most abundant protein of the category is concentrated largely in the Hermes body (figure 4c). All other glycolytic enzymes are characterized by proteomics albeit at markedly different abundances (figure 4d). Thus, the Hermes body concentrates GLUT-3 and all glycolytic enzymes, extending the observations of Yuan et al. [11], who observed only the soluble glycolytic enzymes by their two-dimensional gels.
Figure 4.
Hermes body and glycolysis as deduced from protein abundances and localizations. (a) Twenty-five proteins in metabolism category. Scale indicates percentage total peptides. (b) IF of Hexokinase 1 and (c) IHC of lactate dehydrogenase C. Both show concentration in Hermes bodies of sperm in lumina of the IS. (d) Proteomics of the glycolytic pathway. Histograms represent peptide abundances in separate isolates for the indicated proteins. Total number of peptides (n = 3) characterized in the isolated Hermes bodies is indicated below each protein name. LG, liver Golgi; TG, testis Golgi. Other abbreviations as in figure 2.
3.5. Hermes body concentration of filamentous actin and tubulin for migration
Cytoskeletal proteins are highly abundant (electronic supplementary material, figure S4A) with actin gamma 1 as the most abundant. Tubulin beta 5 and tubulin alpha 1C are also prominent, with the latter localized by LM-IHC to the Hermes body (electronic supplementary material, figure S4b). Myosins (myosin 1B, myosin VB, myosin ID, myosin IIIB) are enriched as well as other prominent cytoskeletal proteins (electronic supplementary material, figure S4a). Several have been localized previously to sperm. These include spectrin [46], BASP1 (NAP22) [47] and the motility-enhancing protein gelsolin [48]. Controls for all LM-IHC or LM-IF using non-immune serum or the absence of primary antibody revealed no immunoreactivity over tissue sections (electronic supplementary material, figure S4C).
Phalloidin staining for filamentous actin visualized its co-localization along with GLUT-3 to the Hermes body (figure 5a–c), as also depicted in transmitted light (figure 5d), merged images (figure 5e), and black and white representations (figure 5f–h). This is the first observation of filamentous actin concentrated in the Hermes body of epididymal sperm.
Figure 5.
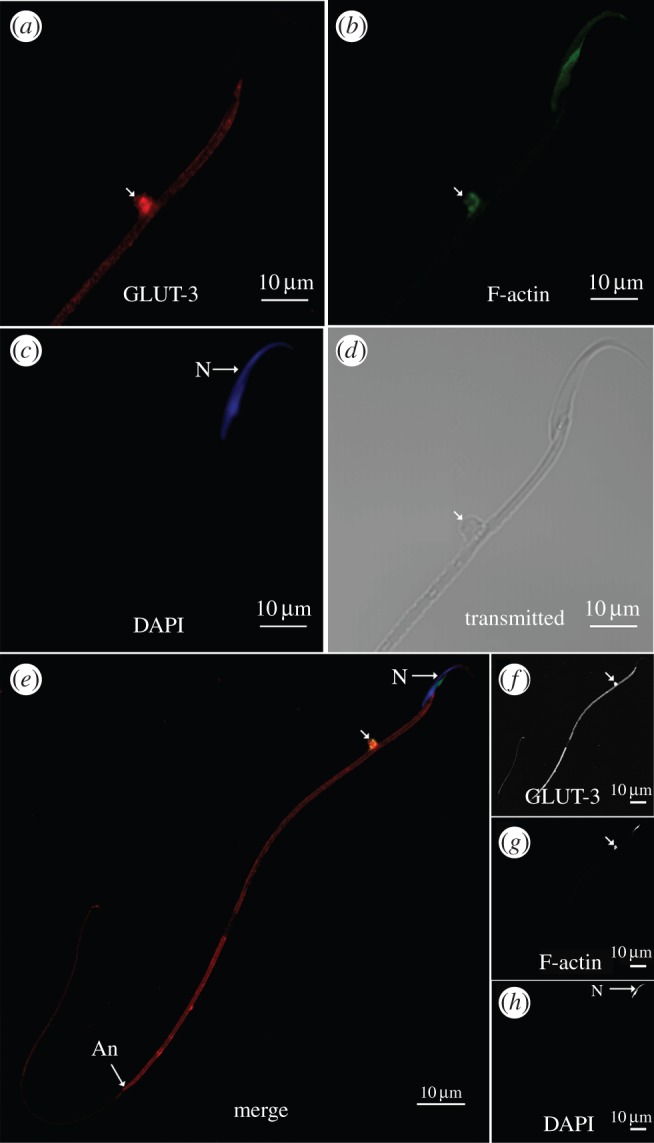
Colocalization of GLUT-3 and filamentous actin in the Hermes bodies of isolated epididymal sperm. (a) Visualization of GLUT-3 and (b) phalloidin (F-actin) in Hermes body. (c) Nucleus (N) stained with DAPI. (d) Sperm head and flagellum in transmitted light. Scale bars = 5 µm. (e) Reactive Hermes body (arrow) with GLUT-3 (red), phalloidin (green) and DAPI (blue) channels merged. Annulus (An). Depicted in black and white are individual reactions for GLUT-3 (f), phalloidin (g) and DAPI (h). Scale bars, 10 µm.
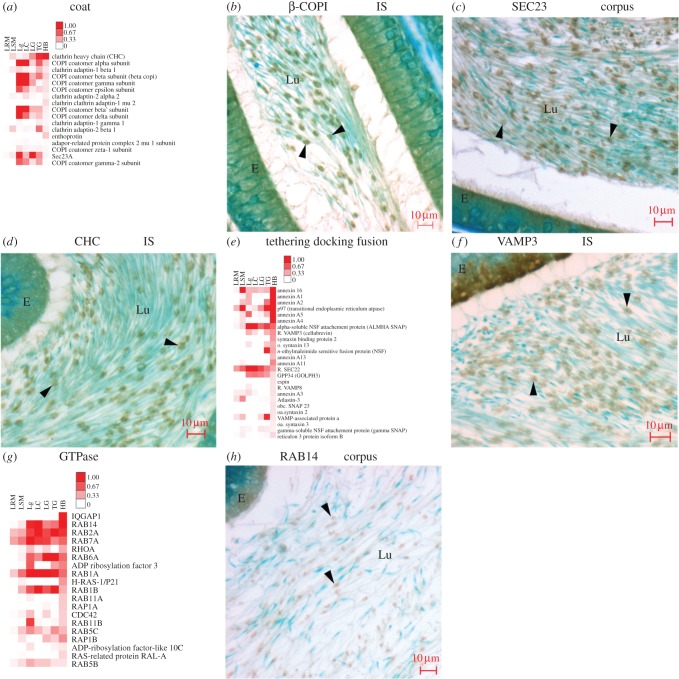
3.6. Extensive membrane trafficking and fusion constituents in Hermes bodies as deduced from abundant COP coats, Clathrin, SNAREs, annexins, AAA+ proteins and trafficking GTPases
COPI and COPII coatomer, clathrin heavy chain and the clathrin adaptins AP1 and AP2 are abundant, as seen in the Coat category (figure 6a). All seven subunits of COPI coatomer of Golgi retrograde directed COPI vesicles [49] are characterized (figure 6a), with β-COP visualized (figure 6b). The anterograde trafficking ER to Golgi vesicles as represented by COPII coatomer subunits are lower in abundance (figure 6a) but readily localized (e.g. Sec23, figure 6c), as were clathrin coat constituents representing clathrin-coated vesicles (e.g. clathrin heavy chain, figure 6d) and its Golgi-restricted adaptor protein AP1 and endocytic-restricted AP2 (not shown). A localization of clathrin heavy chain to Hermes bodies of spermatozoa confirms an earlier study [50].
Figure 6.
Coated vesicle, membrane fusion and Rab proteins in Hermes bodies. (a) Heat map of 17 proteins in coat category sorted by their abundance in Hermes bodies. (b–d) Localization of different coat proteins as indicated for each. (e) Heat map of the most abundant 25 proteins of tethering, docking and fusion sorted by their abundance in Hermes bodies. (f) VAMP3 localization to Hermes bodies. (g) Heat map of the most abundant 19 proteins of GTPase category sorted by their abundance in Hermes bodies. (h) RAB14 localization in Hermes bodies. Abbreviations as in figure 2.
Abundant SNAREs are characterized in the tethering, docking and fusion category (figure 6e; e.g. VAMP3, figure 6f). Annexins are highly abundant, as are the AAA proteins p97 (see below), NSF and its associated alpha SNAP (figure 6e). The atlastins and reticulons, known to be involved in membrane curvature, tubulation and fusion [51], are also abundant in the Hermes body (figure 6e).
The most abundant GTPase is IQGAP (figure 6g), known to coordinate actin polymerization and microtubule formation [52] as well as signalling [53]. The GTPase category includes abundant Rab trafficking proteins. The most abundant Rab is Rab14 (figure 6g), known to be involved in the trafficking and internalization of the GLUT-3 paralogue, GLUT-4, in other cell types [54,55]. Rab14 concentrates in the Hermes body of epididymal sperm in situ (figure 6h). The Golgi marker Rab6a was the sixth most abundant GTPase.
3.7. ER, ribosomal and elongation factor proteins are abundant
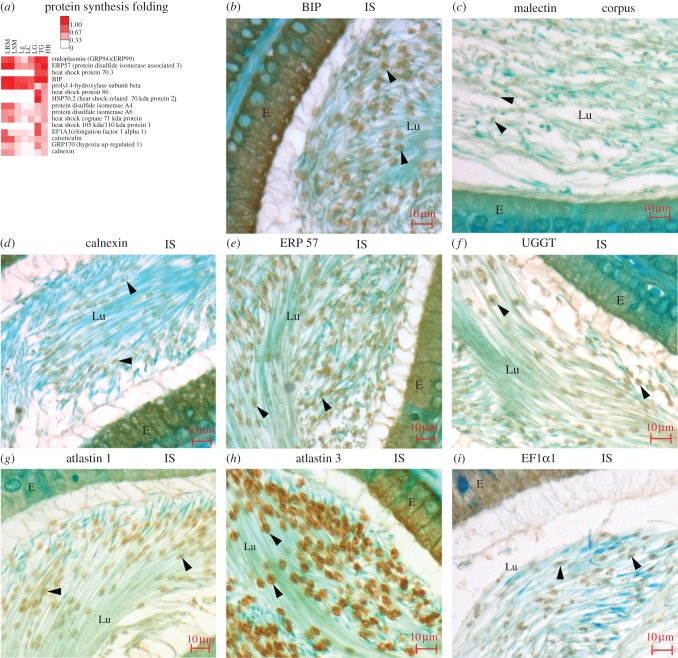
Categorized in the protein synthesis and folding category (figure 7a), several ER proteins concentrate in the Hermes body including BIP, malectin, calnexin, ERP57 and UGGT (figure 7b–f). Proteins involved in ER membrane curvature (reticulons 1 and 3) and fusion (atlastins 1, 3) (figure 7g–h) are also abundant, as indicated in electronic supplementary material, table S2. This is the first localization of these proteins to the Hermes body. As expected from the prior studies of Chauvin et al. [44] on whole epididymal sperm, 68 ribosomal proteins are characterized, as well as the ribosome-associated translation elongation factors EF1A1 and EF2 (electronic supplementary material, table S2), with EF1A2 localized in situ (figure 7i). These data support the hypothesis of Chauvin et al. [44] of an intact translation apparatus in sperm, which we localize to the Hermes body.
Figure 7.
ER proteins are concentrated in isolated Hermes bodies and localized to Hermes bodies in situ. (a) Most abundant 15 proteins of protein synthesis and folding category. (b–i) Localization of abundant ER proteins as indicated. Abbreviations as in figure 2.
3.8. Abundant secretory proteins from Sertoli cells and epithelial lining cells are enriched on Hermes bodies
By proteomics, abundant secretory proteins are observed in the isolated Hermes body (categorized under biosynthetic cargo; electronic supplementary material, figure S5). Albumin, transferrin and ceruloplasmin are highly abundant, but these have previously been considered as serum-derived contaminants of epididymal spermatozoa [44]. Such contamination is ruled out by our proteomic data. ApoE, complement C3 and hemopexin are diminished in the Hermes bodies as compared with liver organelles. These proteins are exclusively made in liver, while albumin and transferrin are biosynthesized in Sertoli cells [56,57]. As expected, albumin and transferrin are localized to the Hermes body in situ (electronic supplementary material, figure S5b–c) as well as in the Sertoli cell of the testis where they are synthesized (electronic supplementary material, figure S5d). Their selective concentration is consequent with their adsorption as Sertoli cell-secreted proteins. Albumin is needed for cholesterol removal and the acquisition of fertility [58,59]. Transferrin and ceruloplasmin are needed for iron uptake [60] as an essential feature for all living cells.
Other proteins (electronic supplementary material, figure S5a), derived from epithelial epididymal cells, also associate with the sperm surface and are seen in the isolated Hermes body fractions (e.g. cysteine-rich secretory protein 1 [61], epididymal-expressed lipocalin-5 [62] and clusterin [63]).
3.9. The Hermes body compartmentalizes stress proteins
By proteomics, several antioxidant enzymes, especially glutathione S transferases (GSTs), are abundant and classified in the detoxification category (electronic supplementary material, figure S6a). Several of these have been localized previously to cytoplasmic droplets [64,65]. However, we can add 18 different heat shock proteins as protective against stress (characterized in the protein synthesis and folding category; figure 7a). This includes HSP70.3 and HSP86 (figure 7a), and the germ cell-specific HSP70.2 protein (localized in electronic supplementary material, figure S6b,c). The HSP70 ER luminal protein, GRP170 (localized in electronic supplementary material, figure S6d), is abundant (figure 7a). The peroxiredoxins 1, 3 and 6, of sperm [66], scavenge reactive oxygen species and are classified in the Metabolism category (figure 4a; localization of peroxiredoxin1 in electronic supplementary material, figure S6e and peroxiredoxin 6 in figure S6f). Our work is the first to reveal a Hermes body localization for several of these proteins that provide protection of whole sperm to oxidative stress [67–69].
3.10. Protein loss in Hermes bodies during sperm transit
Differential protein loss along the epididymal duct is represented by a comparison of two members of the atlastin family of membrane fusion proteins that are evident in the isolated Hermes body (electronic supplementary material, table S2). The gene product expressed in all cells, atlastin 3 [70], and prominent in the Hermes body (figure 6e), is compared with atlastin 1, which is expressed mainly in testis and brain [71]. Atlastin 3 is present throughout the epididymal duct including the cauda region (electronic supplementary material, figure S7a); atlastin 1 is evident in the Hermes body of sperm transiting proximal regions of the duct (electronic supplementary material, figure S7b) but absent from the cauda region (electronic supplementary material, figure S7c). Hence the Hermes body initially expresses both atlastins, but only the ubiquitous atlastin 3 remains in sperm after epididymal release.
As shown in electronic supplementary material, figure S7d, 12 proteins are lost during sperm transit through the epididymis; eight in the caput and four more in the corpus. Consistent with their loss by degradation is the finding of an enrichment of the Hermes body in proteasome constituents and ubiquitin conjugating enzymes categorized in the proteasome category (electronic supplementary material, figure S7e). The most abundant is the known germ cell ubiquitin ligase cullin3 [72], also known to regulate the structure of Golgi apparatus in somatic cells [73]. All ATP-dependent protein degradation constituents, including P97, the 20S proteasome and ubiquitin, are seen in Hermes bodies throughout the epididymal duct (electronic supplementary material, figure S7f–h). Besides revealing the concentration of the regulatory and structural proteins for ATP-dependent proteasome degradation in the Hermes bodies, the selective loss of 12 of the 43 Hermes body proteins during sperm transit in the epididymis supports the hypothesis that ATP-dependent protein degradation in the Hermes body is required for sperm maturation [74,75].
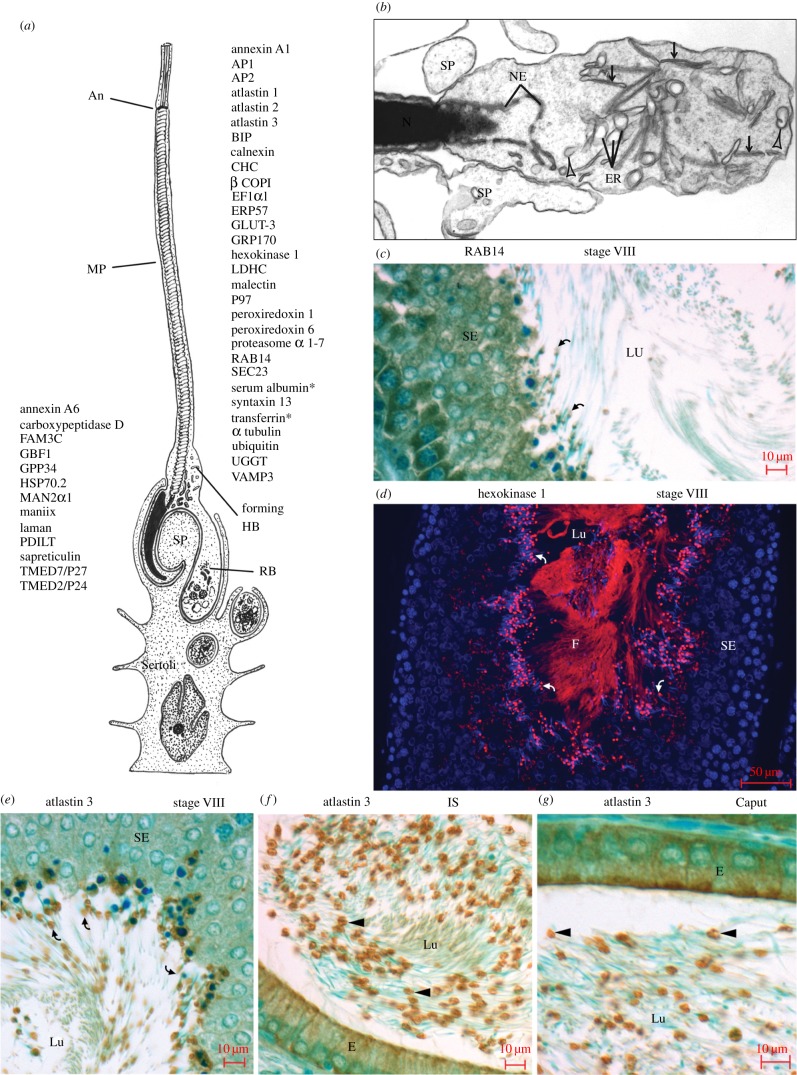
3.11. Hermes body in late step 19 spermatids
The Hermes body forms in late step 19 spermatids as a segregated bulge at the neck or connecting piece of the flagellum (figure 8a), enveloped by Sertoli cell processes (figure 8b). Flattened and dilated cisternae are concentrated in the structure, as are vesicles of different sizes (figure 8b).
Figure 8.
The Hermes body is formed in the step 19 spermatid. (a) Schematic of step 19 spermatid with the flagellar midpiece (MP) ending at the annulus (An) embedded in Sertoli cell processes (SP) that phagocytose excess cytoplasm and organelles into the residual body (RB). Thirteen Golgi-localized proteins of germ cells are indicated on left and 30 non-Golgi proteins on right. All 43 proteins are immunoreactive in step 19 spermatids. Asterisks: Sertoli cell derived albumin and transferrin. (b) EM of step 19 spermatid enveloped by Sertoli cell processes (SP). The forming Hermes body reveals flattened cisternae (arrow), distended membranous elements (DE) and coated vesicles (arrowheads); NE, nuclear envelope; N, nucleus. The forming Hermes bodies (curved arrows) of step 19 spermatids are reactive as noted for Rab14 by IHC (c) and Hexokinase 1 by IF (d) in the SE. (e) Atlastin 3 concentration in the forming Hermes body of step 19 spermatids. (f) Prominent Atlastin 3 immunoreactivity in Hermes bodies of sperm in the IS. (g) Atlastin 3 in Hermes bodies of sperm of the caput epididymidis. Abbreviations as in figure 2.
The hypothesis that the step 19 spermatid concentrated all proteins of the Hermes body of epididymal sperm was tested here by looking at all 43 proteins localized to the Hermes body in situ. Thirteen of these represent Golgi-derived proteins, already demonstrated to be concentrated in the step 19 spermatid by us [14], and summarized on the left of figure 8a. The remaining 30 proteins of Hermes bodies were all localized to the step 19 spermatid in this study (summarized on the right side of figure 8a). Examples are shown for Rab14 (figure 8c) and Hexokinase 1 (figure 8d). As illustrated in figure 8e–g, the abundant smooth ER fusion protein atlastin 3 of the forming Hermes body in step 19 spermatids is compared with that of the Hermes body in epididymal sperm.
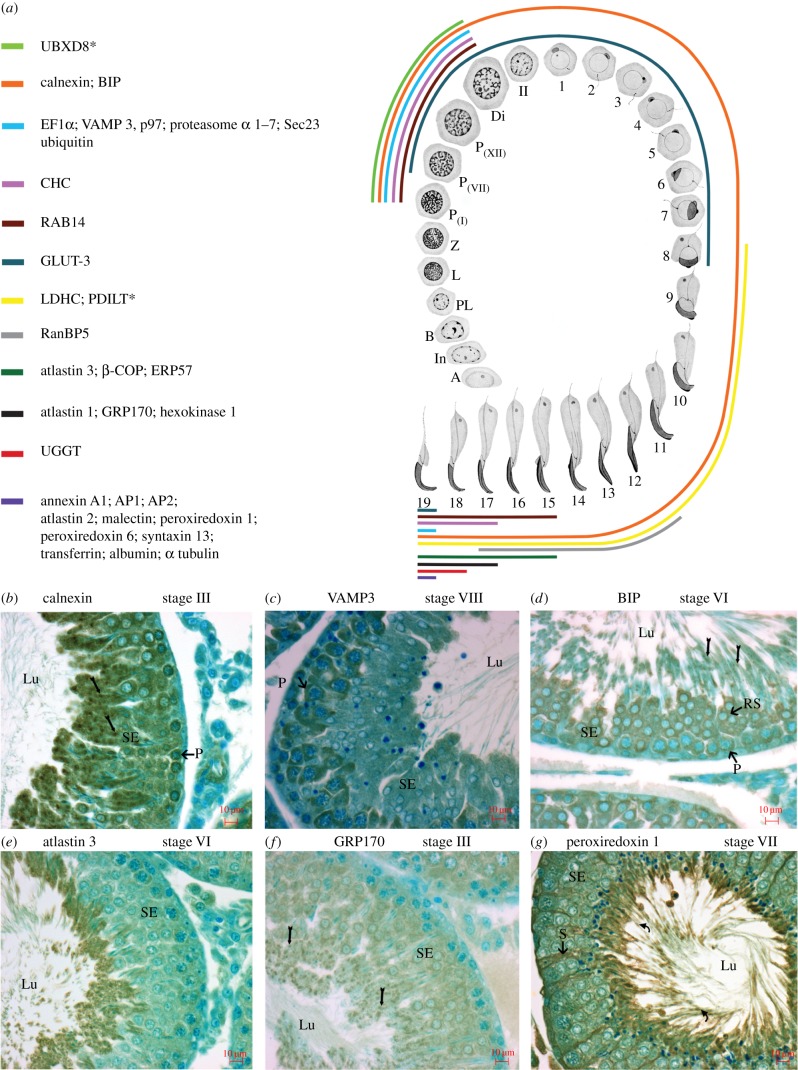
The differential expression of the 30 non-Golgi proteins shown in figure 8a was deduced here for all germ cells during differentiation in the testis (figure 9a), with representative examples shown in figure 9b–g. Taken together with the differential expression of the 13 Golgi proteins previously documented [14], the hypothesis that the step 19 spermatid is the site of Hermes body biogenesis for epididymal sperm is supported at the molecular protein level as well as previously at the morphological level [9].
Figure 9.
Selective protein expression of Hermes body proteins during spermatogenesis. (a) Schematic illustrating distribution of non-Golgi-localized proteins in germ cells during spermatogenesis. Each coloured line represents the expression of immunoreactivity for a non-Golgi-localized protein or combination of proteins in germ cells as they differentiate in the SE of the testis. Spermatogonia (types A, intermediate (In) and B). Primary spermatocytes: preleptotene (PL), leptotene (L), zygotene (Z), pachytene (P), diplotene (Di) and secondary spermatocytes (II), and steps 1–19 of spermiogenesis (modified from [76]). PDILT (asterisk) is also noted as a Golgi-localized protein, as is UBXD8 (asterisk) as previously documented [14]. LM-IHC of the SE of testis immunostained with anti-calnexin (b), Vamp3 (c), BIP (d), atlastin 3 (e), GRP170 (f) and peroxiredoxin 1 (g). Code for labels showing reactivity: pachytene spermatocytes (P), small clumps representative of ER in late spermatids (arrows), forming Hermes bodies (curved arrows), round spermatids (RS), cytoplasmic of late spermatids (asterisks), Sertoli cells (S). Abbreviations as in figure 2.
4. Discussion
4.1. The Hermes body concentrates membrane trafficking proteins, GLUT-3, glycolytic enzymes, filamentous actin and tubulin alpha 1C
This is the first report of abundant membrane trafficking constituents in epididymal sperm here sequestered to the Hermes body. A Golgi apparatus contribution from the TMED proteins is exemplified by TMED7/p27, which localized by EM to the internal flattened cisternae, as did about one-third of GLUT-3, the major plasma membrane protein of epididymal sperm. TMED7/p27 is required to transport plasma membrane cargo from the ER through the Golgi to the plasma membrane [77]. Five TMED paralogues were of high abundance in the Hermes body and together may regulate GLUT-3 expression at the surface as the rate-limiting step in glucose uptake, glycolysis and therefore the initiation of sperm motility.
Besides Golgi contributions, the internal membranes of the Hermes body were deduced as endosomal with clathrin, AP2, VAMP3 and Rab14, indicative of endocytic membrane trafficking [78–80]. Traffic from the ER to the Golgi is exemplified by all constituents of the calnexin cycle, BiP as well as COPII coats for transport to the Golgi. Golgi adaptin AP1 for clathrin-coated vesicles and all COPI coat subunits for Golgi-mediated anterograde COPI vesicle trafficking for intraGolgi and Golgi to ER retrograde traffic are represented, and thus constitute a complete secretory pathway. Also represented are the atlastins and reticulons for ER membrane curvature and fusion.
During epididymal sperm maturation, ATP generation from glycolysis directs the initiation of sperm motility as confined to the principal piece of the sperm flagellum [81–83]. Previous proteomics studies on sperm (including epididymal sperm) [41–43,82] uncovered GLUT-3 and all glycolytic metabolic enzymes as prominent. In humans, 436 proteins of the 1056 found in sperm belong to various metabolic pathways, with glycolytic proteins the highest expressed [84]. The high concentration and localization here of GLUT-3 and glycolytic enzymes to the Hermes body indicates that it is also in this macrostructure that glycolysis generates the ATP needed to initiate sperm motility, and this involves the internal flattened membranes contained therein.
Also shown here for the first time is the sequestration of filamentous actin and the localization of tubulin alpha 1C to the Hermes body in situ in the proximal epididymis. In this region, the Hermes body undergoes a dynamic migration along the flagellar midpiece. This event coincides with the acquisition of sperm motility [5,6,54]. Migration of the Hermes body is coincident with the movement of the internal flattened cisternae in a spiral manner while retaining a close association with the sperm plasma membrane [85]. Prior to the localization of filamentous actin and tubulin alpha 1C here, nothing was known of the cytoskeletal protein constituents that may regulate this migration.
The Hermes body in humans is known to be linked to fertilization [6]. However, the functional significance of the proteins characterized and localized here will require detailed analysis on a protein by protein basis.
4.2. The Hermes body and protein degradation
A complete ATP-dependent protein degradation machinery is concentrated in the Hermes body. This includes ubiquitin, all subunits of the proteasome, P97 and highly abundant cullin 3, the sperm ubiquitin ligase also protective for stress [72,86]. Selective protein loss is shown for 12 proteins of the Hermes body as sperm transit for 10 days from the initial segment to the cauda epididymidis [87]. Proteasome subunits and ubiquitin conjugating enzymes have also been characterized recently in a proteomics study of whole epididymal sperm [44] and localized in situ [88]. Ijiri et al. [89] also observed a loss of proteins during sperm passage through the epididymis. Strikingly, they concluded that HSP70.2 was present in the Hermes body of the caput sperm, but absent from those of the cauda epididymidis exactly as found here. Although Ijiri et al. [89] attributed this to loss of the Hermes body itself, we concluded through the more extensive markers analysed here that the Hermes body is maintained in the cauda epididymidis, although whether or not they are attached to sperm is debatable [2,5]. For humans, the Hermes body appears to be maintained in association with sperm after release from the epididymis [6].
Regardless, selective protein loss through ATP-dependent degradation would modify sperm as they traverse the epididymal duct. Taken together with the proteins of the biosynthetic secretory apparatus characterized here and by Chauvin et al. [44], far from being a cell devoid of a translation machinery, epididymal sperm may be reassessed as in a dynamic state of protein turnover as proposed for all cells [90].
4.3. Hermes body biogenesis at step 19 of spermiogenesis
Transcription ceases immediately after acrosome formation with protein expression thereafter under translational regulation [91]. The translation controlled pathway for organelle modification and morphogenesis of germ cells [92] must be prominent at step 19 to encompass subsets of plasma membrane, cytosolic, Golgi, ER and endosome proteins within the Hermes body of epididymal spermatozoa. That some of these components are merely remnants of the phagocytosis of excess germ cell cytoplasm by Sertoli cells in step 19 spermatids cannot be formally ruled out, but will be resolved as more functional studies ensue.
The work here provides a spatial context for protein concentration in an apparently unique structure designed in epididymal sperm, the Hermes body.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgments
We thank FEMR (http://www.medicine.mcgill.ca/femr/) for EM services (Jeannie Mui) and EM tomography (Kaustuv Basu and Mihnea Bostina). We thank Daniel Friedman, Kristyn Malcolm, Zariyat Mannan, Rebecca Richard, Carl Dahlen, Nadiya Goswami, Abigail Belasen, Maria-Teresa Eyzaguirre, Aurore Fonderflick, Sohee Kang, Dru Perkins, Andrea Prince, Jason Lee, Raja Talla, Rachel Adilman, Krista Dunn, Jamie Koenekoop and Adam Parent for their assistance in LM/IHC. We thank Dr Julie St. Pierre for her help in data interpretation and Hana Hakami, King Saud University, for her help in the manuscript preparation. We are especially grateful to Dr Peter McPherson for atlastin3 antibody as well as the other 19 investigators indicated in the electronic supplementary material, table S3 who generously provided antibodies, advice and guided us to commercial antibodies when available.
Ethics
All animal experiments were carried out in accordance with the Canadian Council on Animal Care and were approved by the McGill University Animal Care Committee (permit no. 4741).
Authors' contributions
C.E.A. and L.H. performed proteomics, subcellular fractionation, imaging and writing of the paper; A.F., subcellular fractionation; C.E.A. and R.E.K., bioinformatics; E.B., J.S., C.E.S., H.V., J.F.-R., P.H.G.S., C.M. and T.N., imaging; J.J.M.B. supervised all aspects and wrote paper with L.H. and C.E.A.
Competing interests
We declare we have no competing interests.
Funding
Supported by CIHR grant MOP 5605. This paper is dedicated to Yves Clermont and C. P. Leblond who elaborated the stem cell renewal theory for male germ cell differentiation. http://www.mcgill.ca/anatomy/stem-cell-renewal-theory.
References
- 1.Retzius G. 1909. Die Spermien der Huftiere. Niol. Untersuch. 14, 163–178. [Google Scholar]
- 2.Hermo L, Dworkin J, Oko R. 1988. Role of epithelial clear cells of the rat epididymis in the disposal of the contents of cytoplasmic droplets detached from spermatozoa. Am. J. Anat. 183, 107–124. (doi:10.1002/aja.1001830202) [DOI] [PubMed] [Google Scholar]
- 3.Fetic S, Yeung CH, Sonntag B, Nieschlag E, Cooper TG. 2006. Relationship of cytoplasmic droplets to motility, migration in mucus, and volume regulation of human spermatozoa. J. Androl. 27, 294–301. (doi:10.2164/jandrol.05122) [DOI] [PubMed] [Google Scholar]
- 4.Rengan AK, Agarwal A, van der Linde M, du Plessis SS. 2012. An investigation of excess residual cytoplasm in human spermatozoa and its distinction from the cytoplasmic droplet. Reprod. Biol. Endocrinol. 10, 92 (doi:10.1186/1477-7827-10-92) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H, Yuan SQ, Zheng ZH, Yan W. 2013. The cytoplasmic droplet may be indicative of sperm motility and normal spermiogenesis. Asian J. Androl. 15, 799–805. (doi:10.1038/aja.2013.69) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper TG. 2011. The epididymis, cytoplasmic droplets and male fertility. Asian J. Androl. 13, 130–138. (doi:10.1038/aja.2010.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orgebin-Crist MC. 1969. Studies on the function of the epididymis. Biol. Reprod. 1(Suppl 1), 155–175. (doi:10.1095/biolreprod1.Supplement_1.155) [DOI] [PubMed] [Google Scholar]
- 8.Cornwall GA. 2009. New insights into epididymal biology and function. Hum. Reprod. Update 15, 213–227. (doi:10.1093/humupd/dmn055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oko R, Hermo L, Chan PT, Fazel A, Bergeron JJ. 1993. The cytoplasmic droplet of rat epididymal spermatozoa contains saccular elements with Golgi characteristics. J. Cell Biol. 123, 809–821. (doi:10.1083/jcb.123.4.809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno RD, Ramalho-Santos J, Chan EK, Wessel GM, Schatten G. 2000. The Golgi apparatus segregates from the lysosomal/acrosomal vesicle during rhesus spermiogenesis: structural alterations. Dev. Biol. 219, 334–349. (doi:10.1006/dbio.2000.9606) [DOI] [PubMed] [Google Scholar]
- 11.Yuan S, Zheng H, Zheng Z, Yan W. 2013. Proteomic analyses reveal a role of cytoplasmic droplets as an energy source during epididymal sperm maturation. PLoS ONE 8, e77466 (doi:10.1371/journal.pone.0077466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng H, Stratton CJ, Morozumi K, Jin J, Yanagimachi R, Yan W. 2007. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc. Natl Acad. Sci. USA 104, 6852–6857. (doi:10.1073/pnas.0701669104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore K, Lovercamp K, Feng D, Antelman J, Sutovsky M, Manandhar G, van Leyen K, Safranski T, Sutovsky P. 2010. Altered epididymal sperm maturation and cytoplasmic droplet migration in subfertile male Alox15 mice. Cell Tissue Res. 340, 569–581. (doi:10.1007/s00441-010-0972-x) [DOI] [PubMed] [Google Scholar]
- 14.Au CE, et al. In press Expression, sorting and segregation of Golgi proteins during germ cell differentiation in the testis. Mol. Biol. Cell. 26 (doi:10.1091/mbc.E14-12-1632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. (doi:10.1006/jsbi.1996.0013) [DOI] [PubMed] [Google Scholar]
- 16.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. (doi:10.1002/jcc.20084) [DOI] [PubMed] [Google Scholar]
- 17.Gilchrist A, et al. 2006. Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281. (doi:10.1016/j.cell.2006.10.036) [DOI] [PubMed] [Google Scholar]
- 18.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJ. 2009. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat. Methods 6, 423–430. (doi:10.1038/nmeth.1333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bencsath-Makkai Z, Bell A, Bergeron J, Boismenu D, Funnell R, Harrison M, Paiement J, Roy L, Kearney R. 2003. Information system for high-throughput proteomics: CellMapBase. Mol. Cell. Proteomics 2, 833. [Google Scholar]
- 20.Bencsath-Makkai Z, et al. 2003. CellMapBase—an information system supporting high-throughput proteomics for the Cell Map Project. IEEE EMBS 4, 3567–3570. (doi:10.1109/IEMBS.2003.1280923) [Google Scholar]
- 21.Kearney R, Blondeau F, McPherson P, Bell A, Servant F, Drapeau M, de Grandpre S, Jm Bergeron J. 2005. Elimination of redundant protein identifications in high throughput proteomics. Conf. Proc. IEEE Eng. Med. Biol. Soc. 5, 4803–4806. (doi:10.1109/IEMBS.2005.1615546) [DOI] [PubMed] [Google Scholar]
- 22.Blondeau F, et al. 2004. Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl Acad. Sci. USA 101, 3833–3838. (doi:10.1073/pnas.0308186101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Sadygov RG, Yates JR III. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201. (doi:10.1021/ac0498563) [DOI] [PubMed] [Google Scholar]
- 24.Li W, Jaroszewski L, Godzik A. 2002. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18, 77–82. (doi:10.1093/bioinformatics/18.1.77) [DOI] [PubMed] [Google Scholar]
- 25.de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20, 1453–1454. (doi:10.1093/bioinformatics/bth078) [DOI] [PubMed] [Google Scholar]
- 26.Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14 863–14 868. (doi:10.1073/pnas.95.25.14863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermo L, Schellenberg M, Liu LY, Dayanandan B, Zhang T, Mandato CA, Smith CE. 2008. Membrane domain specificity in the spatial distribution of aquaporins 5, 7, 9, and 11 in efferent ducts and epididymis of rats. J. Histochem. Cytochem. 56, 1121–1135. (doi:10.1369/jhc.2008.951947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokuyasu KT. 1980. Immunochemistry on ultrathin frozen sections. Histochem. J. 12, 381–403. (doi:10.1007/BF01011956) [DOI] [PubMed] [Google Scholar]
- 29.Zierold K. 1986. The determination of wet weight concentrations of elements in freeze-dried cryosections from biological cells. Scan Electron Microsc. (Pt 2), 713–724. [PubMed] [Google Scholar]
- 30.Simpson IA, Dwyer D, Malide D, Moley KH, Travis A, Vannucci SJ. 2008. The facilitative glucose transporter GLUT3: 20 years of distinction. Am. J. Physiol. Endocrinol. Metab. 295, E242–E253. (doi:10.1152/ajpendo.90388.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujihara Y, Tokuhiro K, Muro Y, Kondoh G, Araki Y, Ikawa M, Okabe M. 2013. Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc. Natl Acad. Sci. USA 110, 8111–8116. (doi:10.1073/pnas.1222166110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yotsumoto H, Sato S, Shibuya M. 1984. Localization of angiotensin converting enzyme (dipeptidyl carboxypeptidase) in swine sperm by immunofluorescence. Life Sci. 35, 1257–1261. (doi:10.1016/0024-3205(84)90096-1) [DOI] [PubMed] [Google Scholar]
- 33.Cherr GN, Yudin AI, Overstreet JW. 2001. The dual functions of GPI-anchored PH-20: hyaluronidase and intracellular signaling. Matrix Biol. 20, 515–525. (doi:10.1016/S0945-053X(01)00171-8) [DOI] [PubMed] [Google Scholar]
- 34.Yoshitake H, Yanagida M, Maruyama M, Takamori K, Hasegawa A, Araki Y. 2011. Molecular characterization and expression of dipeptidase 3, a testis-specific membrane-bound dipeptidase: complex formation with TEX101, a germ-cell-specific antigen in the mouse testis. J. Reprod. Immunol. 90, 202–213. (doi:10.1016/j.jri.2011.04.010) [DOI] [PubMed] [Google Scholar]
- 35.Cardona-Maya W, Velilla PA, Montoya CJ, Cadavid A, Rugeles MT. 2011. In vitro human immunodeficiency virus and sperm cell interaction mediated by the mannose receptor. J. Reprod. Immunol. 92, 1–7. (doi:10.1016/j.jri.2011.09.002) [DOI] [PubMed] [Google Scholar]
- 36.Morales CR, Ni X, Smith CE, Inagaki N, Hermo L. 2012. ABCA17 mediates sterol efflux from mouse spermatozoa plasma membranes. Histol. Histopathol. 27, 317–328. [DOI] [PubMed] [Google Scholar]
- 37.Holm J, Hansen SI, Hoier-Madsen M. 1991. A high-affinity folate binding protein in human semen. Biosci. Rep. 11, 237–242. (doi:10.1007/BF01127499) [DOI] [PubMed] [Google Scholar]
- 38.Jimenez T, McDermott JP, Sanchez G, Blanco G. 2011. Na,K-ATPase alpha4 isoform is essential for sperm fertility. Proc. Natl Acad. Sci. USA 108, 644–649. (doi:10.1073/pnas.1016902108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez T, Sanchez G, McDermott JP, Nguyen AN, Kumar TR, Blanco G. 2011. Increased expression of the Na,K-ATPase alpha4 isoform enhances sperm motility in transgenic mice. Biol. Reprod. 84, 153–161. (doi:10.1095/biolreprod.110.087064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han C, Choi E, Park I, Lee B, Jin S, Kim do H, Nishimura H, Cho C. 2009. Comprehensive analysis of reproductive ADAMs: relationship of ADAM4 and ADAM6 with an ADAM complex required for fertilization in mice. Biol. Reprod. 80, 1001–1008. (doi:10.1095/biolreprod.108.073700) [DOI] [PubMed] [Google Scholar]
- 41.Ikawa M, Tokuhiro K, Yamaguchi R, Benham AM, Tamura T, Wada I, Satouh Y, Inoue N, Okabe M. 2011. Calsperin is a testis-specific chaperone required for sperm fertility. J. Biol. Chem. 286, 5639–5646. (doi:10.1074/jbc.M110.140152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim T, Oh J, Woo JM, Choi E, Im SH, Yoo YJ, Kim DH, Nishimura H, Cho C. 2006. Expression and relationship of male reproductive ADAMs in mouse. Biol. Reprod. 74, 744–750. (doi:10.1095/biolreprod.105.048892) [DOI] [PubMed] [Google Scholar]
- 43.Baker MA, Naumovski N, Hetherington L, Weinberg A, Velkov T, Aitken RJ. 2013. Head and flagella subcompartmental proteomic analysis of human spermatozoa. Proteomics 13, 61–74. (doi:10.1002/pmic.201200350) [DOI] [PubMed] [Google Scholar]
- 44.Chauvin T, Xie F, Liu T, Nicora CD, Yang F, Camp DG II, Smith RD, Roberts KP. 2012. A systematic analysis of a deep mouse epididymal sperm proteome. Biol. Reprod. 87, 141 (doi:10.1095/biolreprod.112.104208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chocu S, Calvel P, Rolland AD, Pineau C. 2012. Spermatogenesis in mammals: proteomic insights. Syst. Biol. Reprod. Med. 58, 179–190. (doi:10.3109/19396368.2012.691943) [DOI] [PubMed] [Google Scholar]
- 46.De Cesaris P, Filippini A, Stefanini M, Ziparo E. 1989. Spectrin, fodrin and protein 4.1-like proteins in differentiating rat germ cells. Differentiation 41, 216–222. (doi:10.1111/j.1432-0436.1989.tb00750.x) [DOI] [PubMed] [Google Scholar]
- 47.Mosevitsky M, Silicheva I. 2011. Subcellular and regional location of ‘brain’ proteins BASP1 and MARCKS in kidney and testis. Acta Histochem. 113, 13–18. (doi:10.1016/j.acthis.2009.07.002) [DOI] [PubMed] [Google Scholar]
- 48.Finkelstein M, Megnagi B, Ickowicz D, Breitbart H. 2013. Regulation of sperm motility by PIP2(4,5) and actin polymerization. Dev. Biol. 381, 62–72. (doi:10.1016/j.ydbio.2013.06.014) [DOI] [PubMed] [Google Scholar]
- 49.Brandizzi F, Barlowe C. 2013. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382–392. (doi:10.1038/nrm3588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Domagala A, Pulido S, Kamieniczna M, Kurpisz M, Herr JC. 2011. An isoimmune response to human sperm clathrin in an infertile woman with systemic lupus erythematosus. J. Reprod. Immunol. 89, 95–102. (doi:10.1016/j.jri.2011.01.015) [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Novick P, Ferro-Novick S. 2013. ER structure and function. Curr. Opin. Cell Biol. 25, 428–433. (doi:10.1016/j.ceb.2013.02.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe T, et al. 2004. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell. 7, 871–883. (doi:10.1016/j.devcel.2004.10.017) [DOI] [PubMed] [Google Scholar]
- 53.Smith JM, Hedman AC, Sacks DB. 2015. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 25, 171–184. (doi:10.1016/j.tcb.2014.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadacca LA, Bruno J, Wen J, Xiong W, McGraw TE. 2013. Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs. Mol. Biol. Cell 24, 2544–2557. (doi:10.1091/mbc.E13-02-0103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stocco DM, Wang X, Jo Y, Manna PR. 2005. Multiple signaling pathways regulating steroidogenesis and steroidogenic acute regulatory protein expression: more complicated than we thought. Mol. Endocrinol. 19, 2647–2659. (doi:10.1210/me.2004-0532) [DOI] [PubMed] [Google Scholar]
- 56.Gelly JL, Richoux JP, Grignon G. 1994. Immunolocalization of albumin and transferrin in germ cells and Sertoli cells during rat gonadal morphogenesis and postnatal development of the testis. Cell Tissue Res. 276, 347–351. (doi:10.1007/BF00306119) [DOI] [PubMed] [Google Scholar]
- 57.Griswold MD. 1988. Protein secretions of Sertoli cells. Int. Rev. Cytol. 110, 133–156. (doi:10.1016/S0074-7696(08)61849-5) [DOI] [PubMed] [Google Scholar]
- 58.Davis BK, Byrne R, Bedigian K. 1980. Studies on the mechanism of capacitation: albumin-mediated changes in plasma membrane lipids during in vitro incubation of rat sperm cells. Proc. Natl Acad. Sci. USA 77, 1546–1550. (doi:10.1073/pnas.77.3.1546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Go KJ, Wolf DP. 1985. Albumin-mediated changes in sperm sterol content during capacitation. Biol. Reprod. 32, 145–153. (doi:10.1095/biolreprod32.1.145) [DOI] [PubMed] [Google Scholar]
- 60.Sylvester SR, Griswold MD. 1994. The testicular iron shuttle: a ‘nurse’ function of the Sertoli cells. J. Androl. 15, 381–385. [PubMed] [Google Scholar]
- 61.Cohen DJ, Da Ros VG, Busso D, Ellerman DA, Maldera JA, Goldweic N, Cuasnicú PS. 2007. Participation of epididymal cysteine-rich secretory proteins in sperm-egg fusion and their potential use for male fertility regulation. Asian J. Androl. 9, 528–532. (doi:10.1111/j.1745-7262.2007.00283.x) [DOI] [PubMed] [Google Scholar]
- 62.Lareyre JJ, Winfrey VP, Kasper S, Ong DE, Matusik RJ, Olson GE, Orgebin-Crist MC. 2001. Gene duplication gives rise to a new 17-kilodalton lipocalin that shows epididymal region-specific expression and testicular factor(s) regulation. Endocrinology 142, 1296–1308. (doi:10.1210/endo.142.3.8045) [DOI] [PubMed] [Google Scholar]
- 63.Hermo L, Wright J, Oko R, Morales CR. 1991. Role of epithelial cells of the male excurrent duct system of the rat in the endocytosis or secretion of sulfated glycoprotein-2 (clusterin). Biol. Reprod. 44, 1113–1131. (doi:10.1095/biolreprod44.6.1113) [DOI] [PubMed] [Google Scholar]
- 64.Papp S, Robaire B, Hermo L. 1995. Immunocytochemical localization of the Ya, Yc, Yb1, and Yb2 subunits of glutathione S-transferases in the testis and epididymis of adult rats. Microsc. Res. Tech. 30, 1–23. (doi:10.1002/jemt.1070300102) [DOI] [PubMed] [Google Scholar]
- 65.Veri JP, Hermo L, Robaire B. 1994. Immunocytochemical localization of glutathione S-transferase Yo subunit in the rat testis and epididymis. J. Androl. 15, 415–434. [PubMed] [Google Scholar]
- 66.O'Flaherty C, de Souza AR. 2011. Hydrogen peroxide modifies human sperm peroxiredoxins in a dose-dependent manner. Biol. Reprod. 84, 238–247. (doi:10.1095/biolreprod.110.085712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aitken RJ, Baker MA. 2006. Oxidative stress, sperm survival and fertility control. Mol. Cell. Endocrinol. 250, 66–69. (doi:10.1016/j.mce.2005.12.026) [DOI] [PubMed] [Google Scholar]
- 68.Aitken RJ, Jones KT, Robertson SA. 2012. Reactive oxygen species and sperm function--in sickness and in health. J. Androl. 33, 1096–1106. (doi:10.2164/jandrol.112.016535) [DOI] [PubMed] [Google Scholar]
- 69.Aitken RJ, Smith TB, Jobling MS, Baker MA, De Iuliis GN. 2014. Oxidative stress and male reproductive health. Asian J. Androl. 16, 31–38. (doi:10.4103/1008-682x.122203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C. 2008. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17, 1591–1604. (doi:10.1093/hmg/ddn046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu PP, Soderblom C, Tao-Cheng JH, Stadler J, Blackstone C. 2006. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum. Mol. Genet. 15, 1343–1353. (doi:10.1093/hmg/ddl054) [DOI] [PubMed] [Google Scholar]
- 72.Wang S, Zheng H, Esaki Y, Kelly F, Yan W. 2006. Cullin3 is a KLHL10-interacting protein preferentially expressed during late spermiogenesis. Biol. Reprod. 74, 102–108. (doi:10.1095/biolreprod.105.045484) [DOI] [PubMed] [Google Scholar]
- 73.Lu A, Pfeffer SR. 2013. Golgi-associated RhoBTB3 targets cyclin E for ubiquitylation and promotes cell cycle progression. J. Cell Biol. 203, 233–250. (doi:10.1083/jcb.201305158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hou CC, Yang WX. 2013. New insights to the ubiquitin-proteasome pathway (UPP) mechanism during spermatogenesis. Mol. Biol. Rep. 40, 3213–3230. (doi:10.1007/s11033-012-2397-y) [DOI] [PubMed] [Google Scholar]
- 75.Sutovsky P. 2011. Sperm proteasome and fertilization. Reproduction 142, 1–14. (doi:10.1530/REP-11-0041) [DOI] [PubMed] [Google Scholar]
- 76.Leblond CP, Clermont Y. 1952. Definition of the stages of the cycle of the seminiferous epithelium in the rat. Ann. N Y Acad. Sci. 55, 548–573. (doi:10.1111/j.1749-6632.1952.tb26576.x) [DOI] [PubMed] [Google Scholar]
- 77.Liaunardy-Jopeace A, Bryant CE, Gay NJ. 2014. The COP II adaptor protein TMED7 is required to initiate and mediate the delivery of TLR4 to the plasma membrane. Sci. Signal. 7, ra70. (doi:10.1126/scisignal.2005275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McMahon HT, Boucrot E. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533. (doi:10.1038/nrm3151) [DOI] [PubMed] [Google Scholar]
- 79.Lim SN, Bonzelius F, Low SH, Wille H, Weimbs T, Herman GA. 2001. Identification of discrete classes of endosome-derived small vesicles as a major cellular pool for recycling membrane proteins. Mol. Biol. Cell 12, 981–995. (doi:10.1091/mbc.12.4.981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reed SE, Hodgson LR, Song S, May MT, Kelly EE, McCaffrey MW, Mastick CC, Verkade P, Tavare JM. 2013. A role for Rab14 in the endocytic trafficking of GLUT4 in 3T3-L1 adipocytes. J. Cell Sci. 126, 1931–1941. (doi:10.1242/jcs.104307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mannowetz N, Wandernoth PM, Wennemuth G. 2012. Glucose is a pH-dependent motor for sperm beat frequency during early activation. PLoS ONE 7, e41030 (doi:10.1371/journal.pone.0041030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miki K. 2007. Energy metabolism and sperm function. Soc. Reprod. Fertility Suppl. 65, 309–325. [PubMed] [Google Scholar]
- 83.Odet F, Gabel S, London RE, Goldberg E, Eddy EM. 2013. Glycolysis and mitochondrial respiration in mouse LDHC-null sperm. Biol. Reprod. 88, 95 (doi:10.1095/biolreprod.113.108530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baker MA, Aitken RJ. 2009. Proteomic insights into spermatozoa: critiques, comments and concerns. Exp. Rev. Proteomics 6, 691–705. (doi:10.1586/epr.09.76) [DOI] [PubMed] [Google Scholar]
- 85.Hermo L, Pelletier RM, Cyr DG, Smith CE. 2010. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 3: developmental changes in spermatid flagellum and cytoplasmic droplet and interaction of sperm with the zona pellucida and egg plasma membrane. Microsc. Res. Tech. 73, 320–363. (doi:10.1002/jemt.20784) [DOI] [PubMed] [Google Scholar]
- 86.Genschik P, Sumara I, Lechner E. 2013. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 32, 2307–2320. (doi:10.1038/emboj.2013.173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robaire B, Hermo L. 1988. Efferent duct, epididymis, and vas defenrens: Structure, functions, and their regulation. In The physiology of reproduction, vol. 1 (eds Knobil E, Neill J), pp. 999–1080. New York, NY: Raven Press. [Google Scholar]
- 88.Fischer KA, Van Leyen K, Lovercamp KW, Manandhar G, Sutovsky M, Feng D, Safranski T, Sutovsky P. 2005. 15-Lipoxygenase is a component of the mammalian sperm cytoplasmic droplet. Reproduction 130, 213–222. (doi:10.1530/rep.1.00646) [DOI] [PubMed] [Google Scholar]
- 89.Ijiri TW, Merdiushev T, Cao W, Gerton GL. 2011. Identification and validation of mouse sperm proteins correlated with epididymal maturation. Proteomics 11, 4047–4062. (doi:10.1002/pmic.201100075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schoenheimer R. 1942. The dynamic state of body constituents. Cambridge, MA: Harvard University Press. [Google Scholar]
- 91.Hecht NB. 1990. Regulation of 'haploid expressed genes’ in male germ cells. J. Reprod. Fertil. 88, 679–693. (doi:10.1530/jrf.0.0880679) [DOI] [PubMed] [Google Scholar]
- 92.Chappell VA, Busada JT, Keiper BD, Geyer CB. 2013. Translational activation of developmental messenger RNAs during neonatal mouse testis development. Biol. Reprod. 89, 61 (doi:10.1095/biolreprod.113.109819) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.