Abstract
Papillary renal cell carcinoma (PRCC) represents the second most common histological subtype of RCC, and comprises 2 subtypes. Prognosis for type 1 PRCC is relatively good, whereas type 2 PRCC is associated with poor clinical outcomes. The aim of the present study was to evaluate the clinicopathological and mutations characteristics of PRCC. Hence, we reported on 13 cases of PRCC analyzed using whole-exome sequencing. Histologically, type 2 PRCC showed a higher nuclear grade and lymphovascular invasion rate versus type 1 PRCC (P < 0.05). Immunostaining revealed type 1 PRCC had higher CK7 and lower Top IIα expression rates (P < 0.05). Whole-exome sequencing data analysis revealed that the mutational statuses of 373 genes (287 missense, 69 silent, 6 nonsense, and 11 synonymous mutations) differed significantly between PRCC and normal renal tissues (P < 0.05). Functional enrichment analysis was used to classify the 287 missense-mutated genes into 11 biological process clusters (comprised of 61 biological processes) and 5 pathways, involved in cell adhesion, microtubule-based movement, the cell cycle, polysaccharide biosynthesis, muscle cell development and differentiation, cell death, and negative regulation. Associated pathways included the ATP-binding cassette transporter, extracellular matrix-receptor interaction, lysosome, complement and coagulation cascades, and glyoxylate and dicarboxylate metabolism pathways. The missense mutation status of 19 genes differed significantly between the groups (P < 0.05), and alterations in the EEF1D, RFNG, GPR142, and RAB37 genes were located in different chromosomal regions in type 1 and 2 PRCC. These mutations may contribute to future studies on pathogenic mechanisms and targeted therapy of PRCC.
Keywords: Papillary renal cell carcinoma, whole-exome sequencing, gene mutation
Introduction
Renal cell carcinoma (RCC) accounts for approximately 90% of all renal malignancies. Papillary RCC (PRCC), the second most common RCC subtype, accounting for approximately 10% of all cases, is a renal parenchyma malignant tumor with papillary or tubulopapillary architecture that presents as type 1 or 2 PRCC; type 1 PRCC is composed of single layered small cell and scanty cytoplasm, type 2 PRCC is characterized by pseudostratified large cells and eosinophilic cytoplasm is a renal parenchyma malignant tumor with papillary or tubulopapillary architecture [1]. Based on the cytologic and histologic features, PRCC can be divided into two subtypes, types 1 and 2 [2]. Type 1 PRCC is generally considered to have a better prognosis than type 2 PRCC, although no consensus regarding the standard treatment for metastatic PRCC exists [3-7].
Molecular genetic studies are highly important in diagnosis and prognosis evaluation, and may provide treatment directions. MET locates at 7q, and its mutation relates to susceptibility to PRCC [8]. Mutations of MET have been identified to cause hereditary PRCC, and occur in a small proportion of sporadic PRCC and a greater number show somatic copy number gains involving chromosome 7q [8,9]. In addition, leucine-rich repeat kinase 2 (LRRK2) is overexpressed and amplified in PRCC. MET and LRRK2 have a synergistic effect during tumor growth via the mTOR and STAT3 pathway [10]. The exome BeadChip can not only identify gene mutations, but also identify diagnostic and therapeutic oncogenes and tumor suppressor genes. Although the pathologic and immunophenotypic of PRCC have been investigated, whole-genome exon sequencing reports are limited. Therefore, we here examined the clinicopathological and gene mutation characteristics of PRCC by a combination of immunohistochemistry and exon chip analyses.
Materials and methods
Specimens
The study contained 13 paraffin-embedded PRCCs and 18 normal kidney tissues. 13 tumors consisted of 6 case of type 1 and 7 case of type 2 PRCC. All tissues were obtained from the archives of the Department of Pathology, School of Medicine, Shihezi University. After asked for the view of the patients and the Institutional Research Ethics Committee, we make a collection of the clinicopathological data for these cases in the patients’ medical records. All specimens were observed by two independent pathologists. Nuclear grading was done according to the Fuhrman nuclear grade system. Tumor stages were according to the 2010 TNM (T = Tumor, N = Node, M = Metastases) classification of the American Joint Committee on Cancer.
Immunohistochemistry (IHC)
IHC staining was performed on 4 μm thick formalin-fixed, paraffin-embedded tissue sections by the 2-step Envision technique (Dako, Denmark). The primary antibodies included cluster of differentiation (CD) 10 (GT200410, 1:100), cytokeratin (CK) (AE1/AE3, 1:100), vimentin (Vim3B4, 1:100), CD117 (1:300), alpha-methylacyl-CoA racemase (AMACR), Top IIα, MDM2, p53, (13H4, 1:100), and CK7 (OV-TL12/30, 1:50), and purchased from Dako company. Negative or positive control was set up on the basis of antibodies.
DNA extraction
Total DNA was isolated from the 13 cases of PRCC and 18 cases of normal kidney tissue samples by using a standard phenol/chloroform extraction method. The quantity of DNA was measured by reading A260/280 ratios by spectrophotometer. When A260/280 ratios located range 1.8 to 2.0, DNA was available. Extractions were stored at -80°C until they were labeled by nick translation.
Whole-exome sequencing
A total of 1 μg of DNA from each of the 13 PRCC tissues and 18 normal kidney tissues were labeled with Illumina reagents and hybridized to Human Exome BeadChips (Illumina, USA). The quality assessment was performed by Illumina Expression Console software. Compared with normal renal tissues, the mutative genes were identified the mutated genes by significance analysis of microarrays (SAM) algorithm in PRCC tissues. The mutative genes associated with cell cycle regulation and other biological functions were determined by Gene Ontology biological process (Gene Ontology BP) enrichment of the classification analysis. The pathways associated with PRCC were confirmed by the Kyoto Encyclopedia of Genes and Genomes database (KEGG).
Statistical analysis
All statistical calculations were done using SPSS 17.0. Difference of measurement data was compared with single factor analysis of variance. Count data were analyzed using Fisher’s exact test. Classification enrichment of gene function and pathway were used to analyze gene function (Gene Ontology of Biological Processes, Molecular function) by DAVID database and KEGG Database. P value < 0.05 was a difference in statistics.
Results
Clinical features
The clinical characteristics of type 1 and type 2 PRCC are summarized in Table 1. In this cohort, 7 patients were men and 6 were women (1.2 male/female ratio); mean age was 53.9 (range from 26 to 74); the average age of the patients was 61.5 (range from 48 to 74) with type 1 PRCC, 47.4 (range from 26 to 63) with type 2 PRCC. The male-to-female ratio, the mean age of the patients, and metastasis were not significantly different between the two groups. In the 13 cases, 3 were asymptomatic, 6 were presented with osphyalgia, and 4 were presented with hematuria. Ultrasonic examination and Computed tomography (CT) showed inhomogeneous mass, as the tumor mass often had hemorrhage, necrosis, or cystic degeneration. Follow-up found the tumor related survival rate was 82.7% (5/6) for the patients with type 1 and 28.6% (2/7) for those with type 2 PRCC. According to 2010 AJCC staging criteria, 6 neoplasms presented at stage 1, 4 at stage 2, 3 at stage 3, 0 at stage 4.
Table 1.
Distribution of analyzed clinicopathologic features and outcome of type 1 and type 2 PRCC
| Characteristics | Type 1 | Type 2 | P value | |
|---|---|---|---|---|
| No. of patients | 6 | 7 | ||
| Age of patients | Mean ± SD | 61.5 ± 11.8 | 47.4 ± 13.7 | 0.075 |
| Range | 48-74 | 26-63 | ||
| Sex of patients | Male | 3 | 4 | |
| Female | 3 | 3 | 0.617 | |
| Metastasis | Positive | 0 | 1 | 0.538 |
| Tumor size (cm) | Mean ± SD | 6.92 ± 3.06 | 7.27 ± 3.10 | 0.840 |
| Range | 3.5-11 | 3.9-13 | ||
| Fuhrman grade | Low (1-2) | 6 | 2 | |
| High (3) | 0 | 5 | 0.049 | |
| Lymphovascular invasion | Negative | 6 | 3 | |
| Positive | 0 | 4 | 0.049 | |
| stage | I-II | 6 | 4 | |
| III -IV | 0 | 3 | 0.122 | |
| Outcome | Dead | 5 (5/6) |
P value: type 1 PRCC vs. type 2 PRCC; Fisher’s exact test.
Histopathology
The differences of histopathology between the two types were described in Table 1. All tumors were located in unilateral renal parenchyma; Mean tumor size, calculated on the maximum diameter, was 7.11 cm (range from 3.5 to 13 cm); it was no difference between the two types. Tumor color are gray, gray yellow, gray red, or colorful; necrosis and hemorrhage could be observed in 4 cases grossly. Microscopically, the tumor was mainly composed of the different proportion of papillary and tubular structure. They were composed of cells arranged on a delicate fibrovascular core. The cytoplasm may be basophilic, eosinophilic, or sometimes partially clear. 6 (6/13) cases were diagnosed as type 1 (Figure 1A) and 7 (7/13) cases were type 2 PRCC (Figure 1B) by their appearance under a microscope. Type 2 PRCC had higher nuclear grade (P = 0.049) and Lymphovascular invasion in relative to type 1 (P = 0.049).
Figure 1.
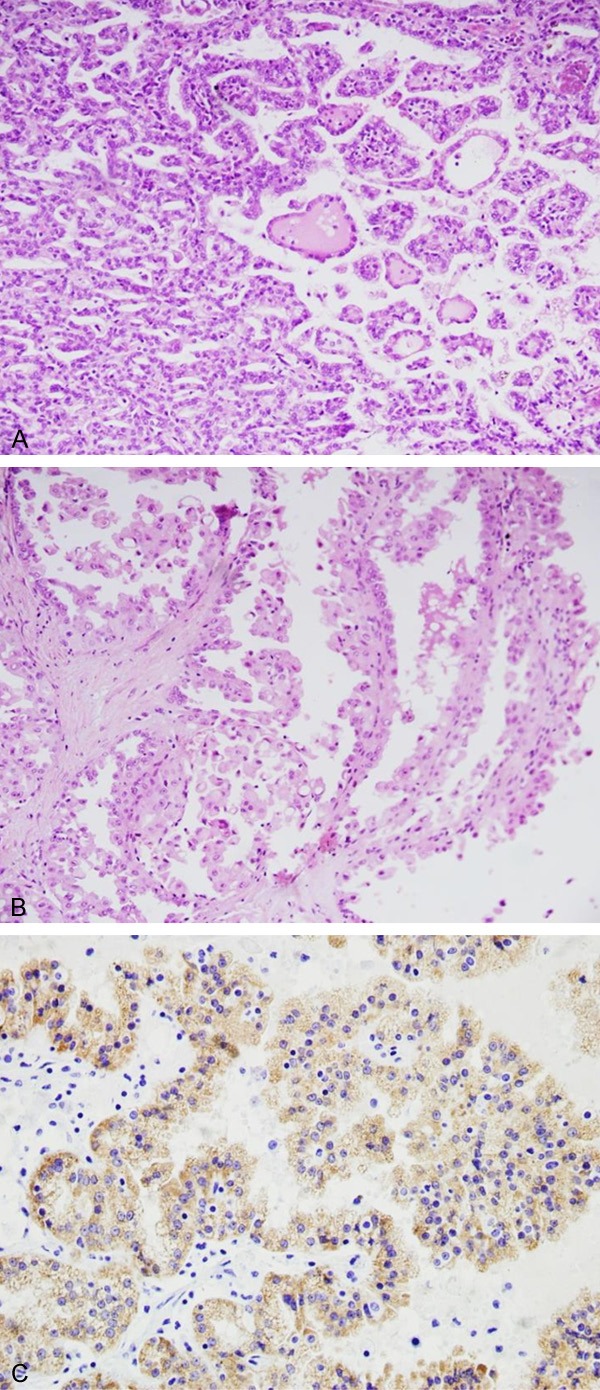
Microscopic and immunohistochemical findings in PRCC. A. Type 1 PRCC was papillae covered by small tumor cells with scanty and basophilic cytoplasm and round nucleus, arranged in a single layer on papillary basement membrane. (H&E, × 200); B. Type 2 PRCC was pseudostratified ciliated columnar epithelium on papillary cores, often with abundant and eosinophilic cytoplasm, large nuclei and prominent nucleoli (H&E, × 200); C. Immunohistochemically, PRCC showed diffuse intense plasma membrane staining for AMACR. (× 200).
Immunohistochemistry
Results of immunohistochemical staining were summarized in Table 2. All PRCC expressed AMACR (Figure 1C), CK7 positive expression rate of type 1 PRCC (6/6) was higher in compared with type 2 (2/7) (P = 0.016). In contrast, Top IIα immunoreactivity was negative (0/6) in type 1 PRCC, while the majority of type 2 PRCC (4/7) were positive for Top IIα (P = 0.049).
Table 2.
Immunohistochemical analyses of CK, CD10, Vimentin, AMACR, CK7, CD117, Top IIα, MDM2, and p53 in PRCC
| Antigen | PRCC | Type 1 | Type 2 | P value |
|---|---|---|---|---|
|
|
||||
| % (n) | % (n) | % (n) | % (n) | |
| CK | 84.6 (11/13) | 83.3 (5/6) | 85.7 (6/7) | |
| CD10 | 30.8 (4/13) | 50 (3/6) | 14.3 (1/7) | |
| Vimentin | 30.8 (4/13) | 33.3 (2/6) | 28.6 (2/7) | |
| AMACR | 100 (13/13) | 100 (6/6) | 100 (7/7) | |
| CK7 | 61.5 (8/13) | 100 (6/6) | 28.6 (2/7) | 0.016 |
| CD117 | 30.8 (4/13) | 33.3 (2/6) | 28.6 (2/7) | |
| Top IIα | 30.8 (4/13) | 0 (0/6) | 57.1 (4/7) | 0.049 |
| MDM2 | 0 (0/13) | 0 (0/6) | 0 (0/7) | |
| P53 | 30.8 (4/13) | 7.7 (1/6) | 42.9 (3/7) | |
P value: type 1 PRCC vs. type 2 PRCC; Fisher’s exact test.
Whole-exome sequencing
In the whole-exome sequencing data analysis, the mutational status of 373 genes was found to be significantly different (P < 0.05) between PRCC and normal renal tissues. In PRCC tissues, 287 missense, 69 silent, 6 nonsense, and 11 synonymous mutations were detected (Table 3). In the functional enrichment analysis, the 287 missense-mutated genes were classified into 11 biological process clusters (comprised of 61 biological progresses) and 5 pathways (P < 0.05) (Table 4; Figure 2A). Mutated genes in PRCC tissues were mainly involved in cell adhesion, microtubule-based movement, cell cycle process, polysaccharide biosynthetic process, tissue morphogenesis, muscle cell development, cell death, differentiation maintenance of organ identity, negative regulation, fertilization, synapsis. Associated pathways included ABC transporters (ATP-binding cassette transporter), ECM (extracellular matrix)-receptor interaction, Lysosome, Complement and coagulation cascades, and Glyoxylate and dicarboxylate metabolism.
Table 3.
The 287 genes containing missense mutations detected in the PRCC tissues (p < 0.05)**
| Chr | SNP_name | Alleles | Gene | p | Mutation(s) |
|---|---|---|---|---|---|
| 1 | exm112317 | [T/C] | CD1C | 0.039215686 | Missense_A118V |
| 1 | exm103938 | [T/C] | UBAP2L | 0.027634131 | Missense_A642V, Missense_A642V |
| 1 | exm134287 | [A/C] | ASPM | 0.049773756 | Missense_A663S, Missense_A663S |
| 1 | exm112233 | [C/G] | CD1A | 0.033333333 | Missense_C68W |
| 1 | exm135632 | [A/G] | CAMSAP2 | 0.032967033 | Missense_D257N |
| 1 | exm124480 | [T/C] | CENPL | 0.045454545 | Missense_D285G, Missense_D285G, Missense_D331G |
| 1 | exm131535 | [A/G] | HMCN1, MIR548F1 | 0.016983017 | Missense_E2893G, Silent |
| 1 | exm113728 | [C/G] | MNDA | 0.018181818 | Missense_E41Q |
| 1 | exm131223 | [A/G] | HMCN1 | 0.022977023 | Missense_E494K |
| 1 | exm127218 | [C/G] | AXDND1 | 0.014705882 | Missense_E991Q |
| 1 | exm140251 | [A/G] | ZC3H11A | 0.032967033 | Missense_G233S |
| 1 | exm100981 | [T/C] | CRNN | 0.047385621 | Missense_G480S |
| 1 | exm131714 | [T/C] | HMCN1, MIR548F1 | 0.047619048 | Missense_H4084Y, Silent |
| 1 | exm121615 | [T/C] | C1orf114 | 0.047619048 | Missense_I37V |
| 1 | exm112921 | [A/G] | OR10X1 | 0.030969031 | Missense_I60T |
| 1 | exm100957 | [T/A] | FLG2 | 0.009960474 | Missense_L168F |
| 1 | exm139949 | [T/C] | OPTC | 0.013931889 | Missense_L268P |
| 1 | exm142351 | [A/G] | LEMD1 | 0.044117647 | Missense_P25S, Missense_P25S, Missense_P25S, Silent, Missense_P25S |
| 1 | exm113801 | [T/C] | MNDA | 0.028571429 | Missense_P403L |
| 1 | exm138568 | [A/C] | LGR6 | 0.045454545 | Missense_P920T, Missense_P868T, Missense_P781T |
| 1 | exm118835 | [T/C] | C1orf111 | 0.047619048 | Missense_R217H |
| 1 | exm131074 | [T/C] | SWT1 | 0.030701754 | Missense_R656C, Missense_R656C |
| 1 | exm101773 | [T/C] | SPRR4 | 0.033333333 | Missense_R8W |
| 1 | exm1164 | [T/C] | AGRN | 0.008333333 | Missense_T1044M |
| 1 | exm127753 | [T/C] | CEP350 | 0.044117647 | Missense_T1131I |
| 1 | exm131226 | [T/C] | HMCN1 | 0.047619048 | Missense_T512I |
| 1 | exm111790 | [A/C] | FCRL3 | 0.027472527 | Missense_V93G |
| 1 | exm118460 | [C/G] | FCRLB | 0.045454545 | Missense_X427S |
| 1 | exm117119 | [A/C] | KLHDC9 | 0.03250774 | Silent, Silent, Missense_S171R, Missense_S171R |
| 12 | exm1053356 | [A/G] | GALNT9 | 0.047619048 | Missense_A152V, Missense_A518V |
| 12 | exm1025298 | [A/C] | CEP290 | 0.029411765 | Missense_D2396Y |
| 12 | exm1006156 | [T/C] | KRT2 | 0.027472527 | Missense_E376K |
| 12 | exm1054454 | [T/C] | GOLGA3 | 0.028571429 | Missense_G644D, Missense_G644D |
| 12 | exm1023832 | [G/C] | OTOGL | 0.010989011 | Missense_H1239D |
| 12 | exm1002135 | [T/G] | CERS5 | 0.030701754 | Missense_I122L |
| 12 | exm1040411 | [T/A] | RBM19 | 0.013986014 | Missense_K351N, Missense_K351N, Missense_K351N |
| 12 | exm1038106 | [T/C] | NAA25 | 0.024242424 | Missense_K876R |
| 12 | exm1049484 | [A/G] | NCOR2 | 0.047619048 | Missense_P2215L, Missense_P2215L, Missense_P2225L |
| 12 | exm1049813 | [A/G] | NCOR2 | 0.032967033 | Missense_P535L, Missense_P535L, Missense_P536L |
| 12 | exm1024575 | [C/G] | TMTC2 | 0.026315789 | Missense_R139G |
| 12 | exm1006431 | [T/C] | KRT77 | 0.045454545 | Missense_R183Q |
| 12 | exm1034626 | [A/G] | USP30 | 0.026315789 | Missense_R206H |
| 12 | exm1042014 | [A/G] | SRRM4 | 0.010989011 | Missense_R223H |
| 12 | exm1036192 | [A/G] | TCHP | 0.047619048 | Missense_R444H, Missense_R444H |
| 12 | exm1037483 | [A/G] | SH2B3 | 0.034965035 | Missense_R566Q |
| 12 | exm1029879 | [A/G] | UTP20 | 0.015151515 | Missense_R869K |
| 12 | exm1003394 | [T/C] | GALNT6 | 0.008333333 | Missense_S32N |
| 12 | exm1054936 | [A/T] | ZNF268 | 0.022222222 | Missense_S383T, Silent, Silent, Silent, Silent, Silent, Silent, Missense_S383T, Missense_S300T |
| 12 | exm1016108 | [A/C] | INHBC | 0.010989011 | Missense_T166P |
| 12 | exm1053798 | [T/C] | POLE | 0.04743083 | Missense_V1512I |
| 12 | exm1020377 | [T/C] | MDM1 | 0.047619048 | Missense_V348I, Missense_V383I |
| 12 | exm1050732 | [A/G] | TMEM132C | 0.004761905 | Missense_V444I |
| 12 | exm1006093 | [T/C] | KRT73 | 0.036521739 | Missense_V61M |
| 12 | exm1013940 | [T/C] | BAZ2A | 0.045454545 | Missense_V950I |
| 12 | exm1000913 | [A/G] | FAM186B | 0.028571429 | Silent, Missense_P822S |
| 12 | exm1004123 | [A/G] | KRT80 | 0.038461538 | Silent, Missense_S445L |
| 12 | exm1042629 | [T/C] | RAB35 | 0.012254902 | Silent, Missense_V155I |
| 12 | exm1026178 | [T/C] | CLLU1OS, CLLU1 | 0.047619048 | Silent, Silent, Silent, Missense_T106M |
| 13 | exm1055958 | [T/C] | N6AMT2 | 0.030701754 | Missense_A140T |
| 13 | exm1060952 | [A/G] | USPL1 | 0.045454545 | Missense_E1010K |
| 13 | exm1074843 | [A/C] | ABCC4 | 0.022222222 | Missense_G187W, Missense_G187W |
| 13 | exm1062037 | [A/C] | BRCA2 | 0.043956044 | Missense_H2074N |
| 13 | exm1065617 | [T/C] | NAA16 | 0.038461538 | Missense_I547T |
| 13 | exm1057999 | [T/C] | PARP4 | 0.027777778 | Missense_I81V |
| 13 | exm1068871 | [A/T] | SETDB2 | 0.01010101 | Missense_K408I, Missense_K396I |
| 13 | exm1065317 | [T/C] | ELF1 | 0.029411765 | Missense_N58S, Missense_N58S |
| 13 | exm1065781 | [A/G] | KIAA0564 | 0.035714286 | Missense_P1173L |
| 13 | exm1070562 | [A/C] | CKAP2 | 0.024242424 | Missense_P127T, Missense_P128T |
| 13 | exm1061900 | [G/C] | BRCA2 | 0.004761905 | Missense_P655R |
| 13 | exm1064366 | [A/G] | FREM2 | 0.022977023 | Missense_R1668H |
| 13 | exm1069663 | [T/C] | WDFY2 | 0.004995005 | Missense_R168W |
| 13 | exm1062043 | [A/G] | BRCA2 | 0.027634131 | Missense_R2108H |
| 13 | exm1075739 | [A/G] | FARP1 | 0.027472527 | Missense_R411Q |
| 13 | exm1078359 | [A/C] | SLC10A2 | 0.027777778 | Missense_S171A |
| 13 | exm1081805 | [A/G] | GRTP1 | 0.038461538 | Missense_T227M |
| 13 | exm1079428 | [C/G] | COL4A1 | 0.044117647 | Missense_V7L |
| 13 | exm1082605 | [A/G] | GAS6 | 0.015151515 | Silent, Missense_S204L |
| 14 | exm1101474 | [A/G] | FRMD6 | 0.032967033 | Missense_A207T, Missense_A207T |
| 14 | exm1102020 | [C/G] | TXNDC16 | 0.047619048 | Missense_A398G, Missense_A403G |
| 14 | exm1107555 | [A/G] | SYNE2 | 0.012383901 | Missense_A6671T, Missense_A6648T, Missense_A305T, Missense_A179T |
| 14 | exm1129474 | [A/G] | CDC42BPB | 0.035714286 | Missense_A983V |
| 14 | exm1098207 | [C/G] | MIA2 | 0.043956044 | Missense_D547H |
| 14 | exm1096768 | [A/C] | FAM177A1 | 0.027777778 | Missense_E64D, Missense_E87D |
| 14 | exm1115955 | [T/A] | MLH3 | 0.021978022 | Missense_F390I, Missense_F390I |
| 14 | exm1083818 | [T/A] | OR4K13 | 0.049773756 | Missense_I270N |
| 14 | exm1099035 | [C/G] | FANCM | 0.038461538 | Missense_L526V |
| 14 | exm1098233 | [T/C] | CTAGE5 | 0.036363636 | Missense_P28S, Missense_P11S, Missense_P11S, Silent, Missense_P40S, Missense_P40S, Missense_P40S |
| 14 | exm1092597 | [G/C] | REC8 | 0.022268908 | Missense_P294R, Missense_P294R |
| 14 | exm1115047 | [A/G] | LTBP2 | 0.045454545 | Missense_P317L |
| 14 | exm1107750 | [T/C] | MTHFD1 | 0.025641026 | Missense_P328L |
| 14 | exm1125535 | [T/C] | BDKRB2 | 0.027777778 | Missense_R14C |
| 14 | exm1094536 | [T/C] | CMA1 | 0.028571429 | Missense_R151K |
| 14 | exm1084549 | [T/C] | TEP1 | 0.045454545 | Missense_R1772Q |
| 14 | exm1102594 | [T/C] | CGRRF1 | 0.040959041 | Missense_R185W |
| 14 | exm1090676 | [T/C] | MYH6 | 0.038461538 | Missense_R204H |
| 14 | exm1098547 | [T/C] | FSCB | 0.030969031 | Missense_R385Q |
| 14 | exm1117477 | [T/C] | POMT2 | 0.036363636 | Missense_R421Q |
| 14 | exm1099301 | [T/C] | MIS18BP1 | 0.038461538 | Missense_R510Q |
| 14 | exm1129681 | [T/C] | EXOC3L4 | 0.043956044 | Missense_R560C |
| 14 | exm1122333 | [T/C] | RIN3 | 0.032967033 | Missense_R79W |
| 14 | exm1091339 | [A/G] | DHRS2 | 0.035714286 | Missense_R7Q, Missense_R7Q |
| 14 | exm1123266 | [G/C] | UNC79 | 0.029411765 | Missense_S1194C |
| 14 | exm1122077 | [C/G] | ATXN3 | 0.012820513 | ############################################## |
| 14 | exm1098992 | [T/C] | FANCM | 0.047619048 | Missense_S175F |
| 14 | exm1120581 | [T/G] | C14orf102 | 0.045454545 | Missense_S35Y |
| 14 | exm1129309 | [T/C] | AMN | 0.047619048 | Missense_S92L |
| 14 | exm1109913 | [A/G] | ZFYVE26 | 0.008791209 | Missense_T2352I |
| 14 | exm1121606 | [A/G] | CATSPERB | 0.045454545 | Missense_T250M |
| 14 | exm1122447 | [T/C] | RIN3 | 0.045454545 | Missense_T638M |
| 14 | exm1104386 | [A/G] | ARID4A | 0.029411765 | Missense_T779A, Missense_T779A, Missense_T779A |
| 14 | exm1100138 | [T/C] | C14orf183 | 0.042105263 | Missense_V263I |
| 14 | exm1134928 | [A/G] | MTA1 | 0.022222222 | Missense_V372I, Missense_V372I |
| 14 | exm1086223 | [A/G] | ZNF219, C14orf176 | 0.028571429 | Silent, Missense_E208K |
| 15 | exm1171367 | [C/G] | TIPIN | 0.034502262 | Missense_A111G |
| 15 | exm1183486 | [G/C] | FSD2 | 0.034965035 | Missense_A129P |
| 15 | exm1156585 | [A/G] | PPIP5K1 | 0.021708683 | Missense_A1372V, Missense_A1374V, Missense_A1374V, Missense_A1399V |
| 15 | exm1169486 | [T/C] | ANKDD1A | 0.045454545 | Missense_A141V |
| 15 | exm1154945 | [G/C] | TTBK2 | 0.021978022 | Missense_A519P |
| 15 | exm1147190 | [T/C] | ATPBD4 | 0.044117647 | Missense_D46N, Missense_D46N |
| 15 | exm1148492 | [T/C] | PLCB2 | 0.011904762 | Missense_E1110K |
| 15 | exm1152491 | [T/C] | SPTBN5 | 0.035714286 | Missense_E2614K |
| 15 | exm1148014 | [A/G] | EIF2AK4 | 0.017857143 | Missense_E556G |
| 15 | exm1194142 | [A/G] | LRRK1 | 0.018181818 | Missense_G1938D |
| 15 | exm1155304 | [T/G] | TMEM62 | 0.017857143 | Missense_G496V |
| 15 | exm1152787 | [T/C] | SPTBN5 | 0.038461538 | Missense_G800E |
| 15 | exm1179035 | [C/G] | C15orf27 | 0.018181818 | Missense_I141M |
| 15 | exm1152778 | [T/C] | SPTBN5 | 0.027777778 | Missense_K879E |
| 15 | exm1178328 | [G/C] | IMP3 | 0.008791209 | Missense_L182V |
| 15 | exm1171637 | [G/C] | ZWILCH | 0.044117647 | Missense_L569V, Silent |
| 15 | exm1190716 | [A/G] | UNC45A | 0.010989011 | Missense_M249I, Missense_M264I |
| 15 | exm1156124 | [A/G] | TP53BP1 | 0.045454545 | Missense_P1341S, Missense_P1341S, Missense_P1336S |
| 15 | exm1146914 | [A/G] | AQR | 0.045454545 | Missense_P1481L |
| 15 | exm1160701 | [T/G] | ATP8B4 | 0.028571429 | Missense_P371H |
| 15 | exm1147780 | [G/C] | FSIP1 | 0.042105263 | Missense_P541A |
| 15 | exm1157351 | [G/C] | CASC4 | 0.041501976 | Missense_Q113H, Missense_Q113H |
| 15 | exm1164516 | [A/T] | MNS1 | 0.047619048 | Missense_Q151L |
| 15 | exm1191113 | [T/C] | VPS33B | 0.015151515 | Missense_R107Q |
| 15 | exm1192792 | [A/G] | LRRC28 | 0.045454545 | Missense_R109H |
| 15 | exm1158689 | [A/G] | DUOX1 | 0.028571429 | Missense_R1481Q, Missense_R1481Q |
| 15 | exm1181151 | [T/C] | ADAMTS7 | 0.045454545 | Missense_R218H |
| 15 | exm1182530 | [A/G] | IL16 | 0.035714286 | Missense_R319H, Missense_R319H |
| 15 | exm1153234 | [A/G] | PLA2G4D | 0.005546956 | Missense_R333W |
| 15 | exm1183163 | [A/G] | FAM154B | 0.047619048 | Missense_R389H |
| 15 | exm1186633 | [A/G] | ACAN | 0.027472527 | Missense_R394Q, Missense_R394Q |
| 15 | exm1173050 | [A/G] | PAQR5 | 0.045454545 | Missense_S11N, Missense_S11N |
| 15 | exm1171891 | [A/G] | AAGAB | 0.003611971 | Missense_S220P |
| 15 | exm1176284 | [A/G] | STRA6 | 0.024242424 | Missense_S58L, Missense_S73L, Missense_S95L, Missense_S58L, Missense_S58L, Missense_S58L, Missense_S97L, Missense_S58L |
| 15 | exm1184631 | [A/G] | ZNF592 | 0.046800826 | Missense_S926N |
| 15 | exm1173024 | [A/G] | GLCE | 0.024509804 | Missense_T453A |
| 15 | exm1148275 | [T/C] | BUB1B | 0.027667984 | Missense_T648I |
| 15 | exm1172534 | [A/G] | ITGA11 | 0.042105263 | Missense_T960I |
| 15 | exm1185057 | [A/G] | SLC28A1 | 0.035714286 | Missense_V189I |
| 15 | exm1188412 | [T/C] | PLIN1 | 0.035714286 | Missense_V272M, Missense_V272M |
| 15 | exm1178993 | [T/A] | FBXO22, FBXO22-AS1 | 0.030701754 | Missense_X404Y, Silent, Silent |
| 15 | exm1153448 | [T/C] | PLA2G4F | 0.008791209 | Silent, Missense_V247M |
| 16 | exm1229096 | [T/C] | KIAA0556 | 0.035714286 | Missense_A1240V |
| 16 | exm1200654 | [T/C] | CACNA1H | 0.044117647 | Missense_A1942V, Missense_A1936V |
| 16 | exm1198320 | [T/C] | WDR24 | 0.047619048 | Missense_A390T |
| 16 | exm1217326 | [T/C] | CIITA | 0.018181818 | Missense_A506V |
| 16 | exm1244992 | [T/C] | CNGB1 | 0.020979021 | Missense_D402N |
| 16 | exm1243334 | [G/C] | CPNE2 | 0.035714286 | Missense_D82E |
| 16 | exm1259888 | [C/G] | KARS | 0.032967033 | Missense_E120Q, Missense_E92Q |
| 16 | exm1216939 | [T/C] | EMP2 | 0.047619048 | Missense_E121K |
| 16 | exm1227991 | [A/G] | AQP8 | 0.045454545 | Missense_E150K |
| 16 | exm1261087 | [A/T] | CENPN | 0.022727273 | Missense_E84D, Missense_E84D, Missense_E84D |
| 16 | exm1234219 | [A/G] | PHKG2 | 0.049122807 | Missense_G86S, Missense_G86S |
| 16 | exm1202974 | [T/C] | CRAMP1L | 0.045454545 | Missense_I1183T |
| 16 | exm1246300 | [A/C] | CDH11 | 0.006993007 | Missense_I433M |
| 16 | exm1256594 | [T/C] | PKD1L3 | 0.029411765 | Missense_K274E |
| 16 | exm1263840 | [A/G] | DNAAF1 | 0.017404938 | Missense_K393R |
| 16 | exm1224564 | [T/C] | ZP2 | 0.013986014 | Missense_M133V |
| 16 | exm1200229 | [A/G] | CACNA1H | 0.043956044 | Missense_M313V, Missense_M313V |
| 16 | exm1267532 | [T/C] | ZNF469 | 0.010989011 | Missense_P1668L |
| 16 | exm1196373 | [A/G] | TMEM8A | 0.015151515 | Missense_P201S |
| 16 | exm1213609 | [A/G] | NMRAL1 | 0.020979021 | Missense_P252L |
| 16 | exm1220429 | [T/C] | ABCC1 | 0.043956044 | Missense_R1066W, Missense_R1007W, Missense_R951W, Missense_R1010W, Missense_R1066W |
| 16 | exm1200424 | [A/G] | CACNA1H | 0.047385621 | Missense_R1069Q, Missense_R1069Q |
| 16 | exm1221023 | [T/C] | XYLT1 | 0.000333 | Missense_R147Q |
| 16 | exm1250942 | [T/C] | DPEP3 | 0.047619048 | Missense_R154K, Missense_R154K |
| 16 | exm1259635 | [G/C] | TMEM231 | 0.024509804 | Missense_R266T, Missense_R237T, Missense_R121T |
| 16 | exm1263012 | [A/G] | MLYCD | 0.035714286 | Missense_R392Q |
| 16 | exm1247857 | [T/C] | KIAA0895L | 0.026923077 | Missense_R459Q |
| 16 | exm1254754 | [A/G] | HYDIN | 0.026923077 | Missense_R4952W |
| 16 | exm1233884 | [A/G] | SRCAP | 0.045454545 | Missense_R966Q |
| 16 | exm1262654 | [G/C] | SDR42E1 | 0.010882822 | Missense_S10T |
| 16 | exm1265329 | [A/C] | CRISPLD2 | 0.043956044 | Missense_S144R |
| 16 | exm1261567 | [T/G] | PKD1L2 | 0.017857143 | Missense_S1665Y |
| 16 | exm1247524 | [A/C] | CES4A | 0.035714286 | Missense_S258R, Missense_S160R, Missense_S164R |
| 16 | exm1251225 | [A/T] | NFATC3 | 0.045454545 | Missense_S269T, Missense_S269T, Missense_S269T |
| 16 | exm1236325 | [A/G] | ITGAX | 0.029411765 | Missense_T123A |
| 16 | exm1243998 | [A/G] | GPR114 | 0.017857143 | Missense_T20A |
| 16 | exm1207604 | [A/C] | CCNF | 0.032967033 | Missense_T327K |
| 16 | exm1247343 | [T/C] | CES2 | 0.012820513 | Missense_T336M, Missense_T336M, Silent |
| 16 | exm1215572 | [A/G] | NAGPA | 0.025641026 | Missense_T465I |
| 16 | exm1268951 | [A/G] | PIEZO1 | 0.045454545 | Missense_T563M |
| 16 | exm1225201 | [T/C] | VWA3A | 0.022242889 | Missense_T657I |
| 16 | exm1256018 | [T/C] | PHLPP2 | 0.045454545 | Missense_V1282I |
| 16 | exm1252046 | [A/G] | CDH3 | 0.032967033 | Missense_V561M |
| 16 | exm1210668 | [A/T] | MEFV | 0.045454545 | Silent, Missense_D424E |
| 16 | exm1199791 | [C/G] | LMF1 | 0.044117647 | Silent, Missense_P562R, Silent |
| 16 | exm1199880 | [T/C] | LMF1 | 0.047619048 | Silent, Missense_R230Q, Silent |
| 17 | exm1301651 | [A/G] | TBC1D28 | 0.026315789 | Missense_A105V |
| 17 | exm1294762 | [T/C] | MYH2 | 0.024242424 | Missense_A1444T, Missense_A1444T |
| 17 | exm1280149 | [A/G] | SPNS3 | 0.047619048 | Missense_A269T |
| 17 | exm1290905 | [T/G] | ALOXE3 | 0.047619048 | Missense_A282D, Missense_A150D |
| 17 | exm1350119 | [A/G] | SLC39A11 | 0.007936508 | Missense_A287V, Missense_A280V |
| 17 | exm1273436 | [T/C] | GEMIN4 | 0.037409701 | Missense_D929N |
| 17 | exm1283034 | [T/C] | ZNF232 | 0.018181818 | Missense_E36K |
| 17 | exm1314605 | [T/C] | SYNRG | 0.018181818 | Missense_E717K, Missense_E717K, Missense_E716K, Missense_E795K, Missense_E717K, Missense_E717K, Missense_E634K |
| 17 | exm1353827 | [G/C] | GGA3 | 0.047619048 | Missense_E97Q, Silent, Missense_E186Q, Missense_E147Q, Missense_E219Q |
| 17 | exm1294342 | [T/C] | MYH4 | 0.040559441 | Missense_G256D |
| 17 | exm1363303 | [A/G] | GAA | 0.047619048 | Missense_G576S, Missense_G576S, Missense_G576S |
| 17 | exm1338149 | [T/C] | TOM1L1 | 0.045454545 | Missense_L348F |
| 17 | exm1349475 | [A/G] | ABCA10 | 0.029411765 | Missense_L663S |
| 17 | exm1361017 | [T/C] | DNAH17 | 0.04743083 | Missense_M1986V |
| 17 | exm1296641 | [T/C] | COX10 | 0.034965035 | Missense_P104L |
| 17 | exm1317248 | [T/C] | ERBB2 | 0.045454545 | Missense_P1177L, Missense_P1207L |
| 17 | exm1365385 | [A/G] | AATK | 0.045454545 | Missense_P1192S, Missense_P1089S |
| 17 | exm1369369 | [A/G] | FASN | 0.027777778 | Missense_P617L |
| 17 | exm1303396 | [A/G] | ALDH3A1 | 0.038461538 | Missense_P79L, Missense_P79L, Missense_P79L |
| 17 | exm1369510 | [T/G] | CCDC57 | 0.040959041 | Missense_Q810K |
| 17 | exm1321241 | [A/C] | KRT31 | 0.021978022 | Missense_R208L |
| 17 | exm1286100 | [A/G] | DVL2 | 0.028571429 | Missense_R237W |
| 17 | exm1342903 | [A/G] | BRIP1 | 0.018181818 | Missense_R264W |
| 17 | exm1327656 | [T/C] | G6PC3 | 0.028571429 | Missense_R274C, Silent, Silent |
| 17 | exm1284328 | [T/C] | WSCD1 | 0.035714286 | Missense_R303W |
| 17 | exm1295950 | [A/G] | DNAH9 | 0.047619048 | Missense_R3726Q, Missense_R38Q |
| 17 | exm1331597 | [A/G] | GOSR2 | 0.046034203 | Missense_R67K, Missense_R67K, Missense_R67K |
| 17 | exm1304297 | [A/G] | KCNJ12, KCNJ18 | 0.040959041 | Missense_R6Q, Missense_R6Q |
| 17 | exm1354940 | [T/C] | RECQL5 | 0.021978022 | Missense_R770Q |
| 17 | exm1311887 | [T/C] | UNC45B | 0.031857032 | Missense_R776W, Missense_R778W |
| 17 | exm1321356 | [A/T] | KRT37 | 0.033333333 | Missense_S73C |
| 17 | exm1295567 | [A/G] | DNAH9 | 0.031620553 | Missense_T1221A |
| 17 | exm1322017 | [A/G] | KRT19 | 0.036119711 | Missense_T327M |
| 17 | exm1337873 | [T/C] | UTP18 | 0.038461538 | Missense_T480I |
| 17 | exm1358363 | [T/C] | AANAT | 0.028571429 | Missense_T76I, Missense_T31I |
| 17 | exm1356970 | [A/G] | EVPL | 0.034965035 | Missense_T835I |
| 17 | exm1326778 | [C/G] | NBR1 | 0.016640867 | Missense_V182L, Missense_V182L, Missense_V182L |
| 17 | exm1364681 | [A/G] | RNF213, LOC100294362 | 0.036363636 | Missense_V4453I, Silent |
| 17 | exm1290479 | [A/G] | GUCY2D | 0.038461538 | Missense_V662M |
| 17 | exm1363356 | [A/G] | GAA | 0.002262443 | Missense_V780I, Missense_V780I, Missense_V780I |
| 17 | exm1345904 | [T/C] | TEX2 | 0.038461538 | Missense_V881M |
| 17 | exm1296979 | [A/T] | FAM18B2-CDRT4, CDRT4 | 0.013986014 | Silent, Silent, Missense_N163Y |
| 17 | exm1288528 | [A/G] | SHBG | 0.043956044 | Silent, Silent, Synonymous_K286K, Missense_D356N, Missense_D241N, Missense_D338N |
| 18 | exm1378989 | [A/G] | LAMA3 | 0.006993007 | Missense_A2146T, Missense_A2090T, Missense_A481T, Missense_A537T |
| 18 | exm1392259 | [A/G] | RTTN | 0.024242424 | Missense_A240V |
| 18 | exm1379174 | [T/C] | TTC39C | 0.024509804 | Missense_A388V, Missense_A449V |
| 18 | exm1378848 | [T/G] | LAMA3 | 0.011904762 | Missense_D1372Y, Missense_D1372Y |
| 18 | exm1371494 | [A/G] | CLUL1 | 0.001262626 | Missense_E173K, Missense_E173K |
| 18 | exm1371832 | [C/G] | METTL4 | 0.045454545 | Missense_E239Q |
| 18 | exm1387358 | [A/G] | SMAD4 | 0.026923077 | Missense_E374K |
| 18 | exm1377011 | [C/G] | MC5R | 0.024242424 | Missense_F209L |
| 18 | exm1381158 | [A/G] | DSG2 | 0.018181818 | Missense_H74R |
| 18 | exm1374780 | [A/G] | ANKRD12 | 0.001262626 | Missense_K906R, Missense_K883R, Missense_K883R |
| 18 | exm1378769 | [T/C] | LAMA3 | 0.031857032 | Missense_L937F, Missense_L937F |
| 18 | exm1378441 | [T/C] | NPC1 | 0.040559441 | Missense_N961S |
| 18 | exm1374337 | [T/C] | CCDC165 | 0.024242424 | Missense_R253W |
| 18 | exm1383557 | [A/G] | TPGS2 | 0.034502262 | Missense_R47C |
| 18 | exm1381499 | [T/C] | TRAPPC8 | 0.044117647 | Missense_R609H |
| 18 | exm1382923 | [A/G] | SLC39A6 | 0.049773756 | Missense_R752C |
| 18 | exm1385694 | [A/G] | KATNAL2 | 0.018181818 | Missense_R85H |
| 18 | exm1381420 | [G/C] | TRAPPC8 | 0.006993007 | Missense_T1223R |
| 18 | exm1382877 | [A/G] | C18orf21 | 0.027472527 | Missense_T44A, Missense_T44A, Silent, Missense_T132A |
| 18 | exm1381205 | [A/G] | DSG2 | 0.027634131 | Missense_V515I |
| 18 | exm1375025 | [A/G] | RALBP1 | 0.030701754 | Missense_V625I |
| 18 | exm1394601 | [C/G] | NFATC1 | 0.019230769 | Silent, Missense_K398N, Missense_K398N, Missense_K385N, Missense_K385N |
| 18 | exm1388566 | [G/C] | LOC100505549, ATP8B1 | 0.020979021 | Silent, Missense_T1242S |
| 19 | exm1420350 | [T/C] | MUC16 | 0.013931889 | Missense_A12925T |
| 19 | exm1422477 | [A/G] | OR7G3 | 0.035714286 | Missense_A237V |
| 19 | exm1404924 | [A/G] | ZNF556 | 0.020639835 | Missense_A248T |
| 19 | exm1396451 | [T/C] | POLRMT | 0.013986014 | Missense_D1085N |
| 19 | exm1427412 | [A/G] | LDLR | 0.036119711 | Missense_D168N, Silent, Missense_D127N, Silent, Missense_D47N, Missense_D168N |
| 19 | exm1401343 | [G/C] | MBD3 | 0.032967033 | Missense_E275D |
| 19 | exm1426542 | [G/C] | SLC44A2 | 0.024509804 | Missense_E550Q, Missense_E552Q |
| 19 | exm1430165 | [C/G] | ZNF20, ZNF625-ZNF20 | 0.036119711 | Missense_F292L, Silent, Missense_F289L |
| 19 | exm1427443 | [A/G] | LDLR | 0.033333333 | Missense_G324S, Missense_G156S, Missense_G283S, Missense_G197S, Missense_G203S, Missense_G324S |
| 19 | exm1422969 | [T/A] | ZNF559-ZNF177, ZNF177 | 0.029411765 | Missense_I295F, Silent, Missense_I295F, Missense_I455F |
| 19 | exm1415176 | [A/G] | EMR1 | 0.044117647 | Missense_I487V, Missense_I398V, Missense_I539V, Missense_I539V, Missense_I362V |
| 19 | exm1422878 | [A/T] | ZNF559, ZNF559-ZNF177 | 0.027568922 | Missense_N364I,Silent, Missense_N258I, Silent, Silent, Missense_N300I, Silent, Silent, Silent, Silent |
| 19 | exm1428643 | [T/G] | RGL3 | 0.049113876 | Missense_P162H, Missense_P162H |
| 19 | exm1397678 | [T/C] | ELANE | 0.033333333 | Missense_P257L |
| 19 | exm1403544 | [A/G] | JSRP1 | 0.047619048 | Missense_P267S |
| 19 | exm1414494 | [T/G] | C3 | 0.018181818 | Missense_P836T |
| 19 | exm1409449 | [A/C] | PLIN4 | 0.032967033 | Missense_R1208M |
| 19 | exm1408191 | [A/G] | CREB3L3 | 0.04747162 | Missense_R392Q |
| 19 | exm1409319 | [T/C] | HDGFRP2 | 0.032967033 | Missense_T50I, Missense_T50I |
| 19 | exm1423686 | [T/C] | COL5A3 | 0.015151515 | Missense_V1691I |
| 19 | exm1415181 | [A/G] | EMR1 | 0.014705882 | Missense_V537I, Missense_V448I, Missense_V589I, Missense_V589I, Missense_V412I |
| 19 | exm1415213 | [G/C] | EMR1 | 0.021708683 | Missense_V672L, Missense_V583L, Missense_V659L, Missense_V724L, Missense_V547L |
| 1 | exm101053 | [T/C] | LCE5A | 0.034965035 | Nonsense_R79X |
| 12 | exm1022625 | [G/C] | GLIPR1L2 | 0.027472527 | Nonsense_Y144X |
| 13 | exm1082947 | [T/G] | UPF3A | 0.038461538 | Nonsense_E258X, Nonsense_E291X |
| 15 | exm1177922 | [A/G] | MAN2C1 | 0.045454545 | Nonsense_R878X, Nonsense_R878X, Nonsense_R895X, Nonsense_R779X |
| 17 | exm1312366 | [A/G] | SLFN13 | 0.049773756 | Nonsense_R647X |
| 17 | exm1352015 | [A/C] | C17orf77 | 0.044117647 | Nonsense_C207X |
| 1 | exm-rs984222 | [C/G] | TBX15 | 0.020979021 | Silent |
| 1 | exm-rs1342038 | [A/G] | LOC100506023 | 0.024509804 | Silent |
| 1 | exm-rs1023252 | [T/G] | CLCN6 | 0.025641026 | Silent |
| 2 | exm-rs1351164 | [T/C] | DIRC3 | 0.025641026 | Silent |
| 2 | exm-rs6732434 | [A/G] | PPP1R1C | 0.035714286 | Silent |
| 2 | exm-rs17027258 | [A/G] | SLC9A4 | 0.043601651 | Silent |
| 2 | exm-rs41464348 | [A/G] | LTBP1 | 0.013986014 | Silent, Silent, Silent, Silent, Silent |
| 3 | exm-rs6439334 | [A/G] | CPNE4 | 0.045454545 | Silent |
| 3 | exm-rs4370013 | [A/T] | CNTN4 | 0.019766611 | Silent, Silent |
| 3 | exm-rs10935120 | [A/G] | CEP63 | 0.010989011 | Silent, Silent, Silent, Silent |
| 4 | exm-rs2273 | [T/C] | SDAD1 | 0.033333333 | Silent |
| 4 | exm-rs1391099 | [A/G] | INPP4B | 0.049773756 | Silent, Silent |
| 5 | exm-rs26232 | [T/C] | C5orf30 | 0.043956044 | Silent |
| 5 | exm-rs31489 | [A/C] | CLPTM1L | 0.044547644 | Silent |
| 6 | exm-rs9276431 | [T/C] | HLA-DQA2 | 0.004545455 | Silent |
| 6 | exm-rs2071556 | [T/G] | HLA-DMB | 0.014705882 | Silent |
| 6 | exm-rs887466 | [A/G] | PSORS1C3 | 0.015151515 | Silent |
| 6 | exm-rs2213568 | [A/C] | HLA-DQA2 | 0.027777778 | Silent |
| 6 | exm-rs2074470 | [A/G] | OR11A1 | 0.028571429 | Silent |
| 6 | exm-rs2242668 | [T/C] | LSM2 | 0.028571429 | Silent |
| 6 | exm-rs6908425 | [T/C] | CDKAL1 | 0.038461538 | Silent |
| 6 | exm-rs444921 | [T/C] | SKIV2L | 0.047385621 | Silent |
| 6 | exm-rs2844775 | [A/G] | TRIM26 | 0.006993007 | Silent, Silent |
| 6 | exm-rs3130383 | [A/C] | TRIM26 | 0.008333333 | Silent, Silent |
| 6 | exm-rs8321 | [A/C] | ZNRD1 | 0.020979021 | Silent, Silent |
| 6 | exm-rs9262113 | [A/G] | PRR3 | 0.027472527 | Silent, Silent |
| 6 | exm-rs9487094 | [A/G] | PPIL6 | 0.027472527 | Silent, Silent |
| 6 | exm-rs3132672 | [A/C] | TRIM26 | 0.027777778 | Silent, Silent |
| 6 | exm-rs4148871 | [A/G] | TAP2 | 0.040090344 | Silent, Silent |
| 6 | exm-rs6916921 | [T/C] | NFKBIL1 | 0.026923077 | Silent, Silent, Silent, Silent |
| 6 | exm-rs707939 | [A/C] | MSH5, MSH5-SAPCD1 | 0.004662005 | Silent, Silent, Silent, Silent, Silent |
| 6 | exm-rs620202 | [T/G] | BRD2 | 0.027472527 | Silent, Silent, Silent, Silent, Silent |
| 6 | exm-rs485502 | [T/C] | BRD2 | 0.035714286 | Silent, Silent, Silent, Silent, Silent |
| 7 | exm-rs730497 | [A/G] | GCK | 0.012820513 | Silent |
| 7 | exm-rs864745 | [T/C] | JAZF1 | 0.049773756 | Silent |
| 7 | exm-rs10256972 | [A/C] | C7orf50 | 0.043956044 | Silent, Silent, Silent |
| 8 | exm-rs7009183 | [A/G] | LOC100616530 | 0.042986425 | Silent, Silent,S ilent, Silent, Silent, Silent, Silent, Silent, Silent |
| 9 | exm-rs10491539 | [T/G] | SH3GL2 | 0.011363636 | Silent |
| 9 | exm-rs17584499 | [T/C] | PTPRD | 0.047619048 | Silent |
| 10 | exm-rs7085433 | [A/G] | TIMM23 | 0.031991744 | Silent |
| 10 | exm-rs1913517 | [A/G] | WDFY4, LRRC18 | 0.04747162 | Silent, Silent |
| 11 | exm-rs7926971 | [A/G] | TEAD1 | 0.028101929 | Silent |
| 12 | exm-rs2970818 | [A/T] | C12orf4 | 0.018059856 | Silent |
| 12 | exm-rs10846934 | [T/C] | TMEM132B | 0.018942963 | Silent |
| 12 | exm-rs1491942 | [G/C] | LRRK2 | 0.027472527 | Silent |
| 12 | exm-rs12579350 | [A/G] | ANO2 | 0.035947712 | Silent |
| 12 | exm-rs7134594 | [T/C] | MMAB | 0.040559441 | Silent, Silent |
| 12 | exm1036101 | [T/G] | TCHP | 0.045454545 | Silent, Silent |
| 14 | exm-rs7140150 | [T/C] | FRMD6 | 0.019230769 | Silent |
| 14 | exm-rs7159841 | [T/C] | MDGA2 | 0.033841159 | Silent |
| 15 | exm-rs4775785 | [T/C] | SHC4 | 0.047619048 | Silent |
| 15 | exm-rs6494537 | [T/C] | DENND4A | 0.034965035 | Silent, Silent |
| 15 | exm-rs12440440 | [A/G] | RYR3 | 0.042744021 | Silent, Silent |
| 15 | exm-rs1378942 | [A/C] | CSK | 0.044117647 | Silent, Silent |
| 15 | exm-rs886144 | [T/C] | SV2B | 0.044117647 | Silent, Silent |
| 15 | exm-rs12915189 | [A/G] | CRTC3 | 0.047619048 | Silent, Silent |
| 15 | exm-rs8043440 | [T/C] | GABRB3 | 0.047619048 | Silent, Silent |
| 16 | exm-rs6564869 | [A/C] | GAN | 0.035714286 | Silent |
| 17 | exm-rs2589133 | [A/G] | RPTOR | 0.027472527 | Silent, Silent |
| 19 | exm1421853 | [T/C] | MUC16 | 0.032967033 | Silent |
| 19 | exm1431071 | [C/G] | MAN2B1 | 0.035714286 | Silent, Silent |
| 19 | exm-rs2279008 | [T/C] | MYO9B | 0.044117647 | Silent, Silent |
| 20 | exm-rs487656 | [A/G] | LOC284757 | 0.043601651 | Silent |
| 21 | exm-rs2826891 | [T/C] | NCAM2 | 0.001998002 | Silent |
| 21 | exm-rs7275212 | [A/T] | ERG | 0.045454545 | Silent, Silent, Silent, Silent, Silent, Silent, Silent |
| 22 | exm-rs139553 | [T/C] | MEI1 | 0.045796309 | Silent |
| Y | exm-rs9341313 | [T/G] | EIF1AY | 0.018181818 | Silent |
| 1 | exm131743 | [T/C] | HMCN1, MIR548F1 | 0.031991744 | Synonymous_A4302A, Silent |
| 1 | exm-rs1142287 | [T/C] | SCAMP3 | 0.032868733 | Synonymous_G126G, Synonymous_G100G |
| 11 | exm-rs4453265 | [T/C] | C2CD3 | 0.047385621 | Synonymous_V1641V |
| 12 | exm1041028 | [T/C] | RNFT2 | 0.017857143 | Synonymous_L183L, Synonymous_L183L |
| 12 | exm1043795 | [T/C] | ACADS | 0.043956044 | Synonymous_N120N |
| 12 | exm1051188 | [T/C] | PIWIL1 | 0.034965035 | Synonymous_A26A, Synonymous_A26A |
| 13 | exm1068030 | [A/G] | ESD | 0.015151515 | Synonymous_I157I |
| 16 | exm1215196 | [A/G] | PPL | 0.038461538 | Synonymous_A826A |
| 16 | exm1244602 | [T/C] | KATNB1 | 0.033333333 | Synonymous_D410D |
| 16 | exm1210664 | [T/C] | MEFV | 0.032967033 | Synonymous_L590L, Missense_D438N |
| 17 | exm1279218 | [A/G] | ATP2A3 | 0.031857032 | Synonymous_A632A, Synonymous_A632A, Synonymous_A632A, Synonymous_A632A, Synonymous_A632A, Synonymous_A632A, Synonymous_A632A |
PRCC vs. normalrenal tissue; Fisher’s exact test.
Abbreviations: PRCC, Papillaryrenal cell carcinoma; Chr, chromosome; SNP, single nucleotidepolymorphism.
Table 4.
Gene Ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the 287 missense mutated genes of Papillary renal cell carcinoma (p < 0.05) detected by Human Exome BeadChip technology
| Category | Cluster | Term | Gene Count | % | P Value | Genes |
|---|---|---|---|---|---|---|
| GOTERM_BP_FAT | Cell adhesion | GO:0007155~cell adhesion | 65 | 6.220095694 | 1.78E-05 | MTSS1, AEBP1, NRP1, CLSTN2, CLSTN1, PCDHA1, PCDHGA1, CLEC4A, COL12A1, SPON1, RET, PCDHB5, CNTNAP5, CDHR1, COL22A1, MYH9, SSPO, GPR98, SIGLEC1, SIGLEC6, HAS1, CPXM1, RELN, CNTN4, COL24A1, DST, ADAMTS13, ITGA11, CRNN, CDH3, DCHS1, ITGAM, LAMB3, COL7A1, SORBS1, AGGF1, ITGAX, FAT4, FAT1, COL27A1, ACAN, COL6A1, LPP, PPFIBP1, HSPG2, COL15A1, PCDH15, NID1, COL5A3, COL4A6, VWF, CASS4, COL19A1, LAMA3, DSG2, EMR1, ERBB2IP, CDH17, FREM2, TMEM8A, LAMA5, FREM1, MUC5B, CDH11, MUC16 |
| GOTERM_BP_FAT | GO:0022610~biological adhesion | 65 | 6.220095694 | 1.90E-05 | MTSS1, AEBP1, NRP1, CLSTN2, CLSTN1, PCDHA1, PCDHGA1, CLEC4A, COL12A1, SPON1, RET, PCDHB5, CNTNAP5, CDHR1, COL22A1, MYH9, SSPO, GPR98, SIGLEC1, SIGLEC6, HAS1, CPXM1, RELN, CNTN4, COL24A1, DST, ADAMTS13, ITGA11, CRNN, CDH3, DCHS1, ITGAM, LAMB3, COL7A1, SORBS1, AGGF1, ITGAX, FAT4, FAT1, COL27A1, ACAN, COL6A1, LPP, PPFIBP1, HSPG2, COL15A1, PCDH15, NID1, COL5A3, COL4A6, VWF, CASS4, COL19A1, LAMA3, DSG2, EMR1, ERBB2IP, CDH17, FREM2, TMEM8A, LAMA5, FREM1, MUC5B, CDH11, MUC16 | |
| GOTERM_BP_FAT | GO:0007156~homophilic cell adhesion | 16 | 1.531100478 | 0.004375914 | RET, PCDHB5, CLSTN2, CDHR1, CLSTN1, PCDH15, PCDHA1, CDH3, DCHS1, PCDHGA1, DSG2, FAT4, FREM2, CDH17, FAT1, CDH11 | |
| GOTERM_BP_FAT | GO:0016337~cell-cell adhesion | 24 | 2.296650718 | 0.024451853 | RET, CLSTN2, PCDHB5, CDHR1, CLSTN1, PCDH15, CRNN, PCDHA1, MYH9, CDH3, ITGAM, DCHS1, GPR98, PCDHGA1, SIGLEC1, COL19A1, DSG2, FAT4, CDH17, FREM2, FAT1, ACAN, CNTN4, CDH11 | |
| GOTERM_BP_FAT | Microtubule-based movement | GO:0007018~microtubule-based movement | 14 | 1.339712919 | 0.007391122 | DNAH11, DNAH9, OPA1, DNAH12, KIF11, DNAH17, BICD2, DNAH5, KIF2C, MACF1, TUBAL3, DYNC2H1, TUBB1, DST |
| GOTERM_BP_FAT | GO:0030705~cytoskeleton-dependent intracellular transport | 8 | 0.765550239 | 0.020201062 | OPA1, MACF1, MYH2, MYH4, MYH14, MYH6, MYH9, DST | |
| GOTERM_BP_FAT | GO:0000226~microtubule cytoskeleton organization | 15 | 1.435406699 | 0.026238636 | RET, CEP120, KIF11, CETN3, TBCE, BRCA2, PLK1S1, MYH9, KIF2C, SASS6, MACF1, BUB1B, MAP7, TUBB1, DST | |
| GOTERM_BP_FAT | GO:0007010~cytoskeleton organization | 35 | 3.349282297 | 0.018418181 | MTSS1, BMP10, CEP120, CETN3, TTN, KIF2C, MACF1, SORBS1, OBSL1, TUBB1, RET, KIF11, SPTBN5, TBCE, BRCA2, CECR2, ARHGEF17, MYH6, PLK1S1, RICTOR, MYH9, PALLD, PLCE1, SASS6, KRT19, ERBB2IP, XIRP2, LAMA5, LIMCH1, PRR5-ARHGAP8, BUB1B, MAP7, ARAP3, DST, CDC42BPB | |
| GOTERM_BP_FAT | GO:0007017~microtubule-based process | 25 | 2.392344498 | 0.004789572 | DNAH11, DNAH9, DNAH12, CEP120, DNAH17, CETN3, DNAH5, KIF2C, MACF1, DYNC2H1, TUBB1, RET, KIF11, OPA1, TBCE, BRCA2, PLK1S1, MYH9, BICD2, SASS6, TUBAL3, BUB1B, MAP7, DST, MAP3K11 | |
| GOTERM_BP_FAT | Signaling pathway | GO:0007229~integrin-mediated signaling pathway | 11 | 1.052631579 | 0.003945816 | ADAMTS7, VAV3, ITGAX, ERBB2IP, ADAMTS13, LAMA5, ITGA11, MYH9, DST, ITGAM, ADAMDEC1 |
| GOTERM_BP_FAT | GO:0035023~regulation of Rho protein signal transduction | 15 | 1.435406699 | 7.82E-04 | OBSCN, VAV3, RALBP1, PREX2, ARHGEF17, RICTOR, TTN, FARP1, MCF2L2, PLEKHG2, SYDE2, RASGRF2, TIAM1, ARAP3, KALRN | |
| GOTERM_BP_FAT | GO:0051056~regulation of small GTPase mediated signal transduction | 25 | 2.392344498 | 0.004617191 | ERBB2, PREX2, RGL3, TTN, MCF2L2, PLEKHG2, TIAM1, KNDC1, OBSCN, VAV3, RALBP1, SIPA1L2, ARHGEF17, RICTOR, FARP1, PLCE1, SYDE2, RASGRF2, TBC1D28, GRTP1, C6ORF170, RELN, ARAP3, TBC1D8B, KALRN | |
| GOTERM_BP_FAT | GO:0046578~regulation of Ras protein signal transduction | 21 | 2.009569378 | 0.009082897 | OBSCN, VAV3, RALBP1, ERBB2, PREX2, ARHGEF17, RICTOR, TTN, FARP1, MCF2L2, PLCE1, PLEKHG2, SYDE2, RASGRF2, TBC1D28, TIAM1, GRTP1, C6ORF170, ARAP3, TBC1D8B, KALRN | |
| GOTERM_BP_FAT | GO:0007167~enzyme linked receptor protein signaling pathway | 27 | 2.583732057 | 0.046239768 | MTSS1, BMP10, FGFR4, NRP1, LTBP2, ERBB2, BDKRB2, TGFB1, SORBS1, TIAM1, GDF9, AGRN, EGF, ROS1, RET, PTPRG, SMAD4, GUCY2C, EPHA1, EPHA3, GUCY2D, EPHA5, PLCE1, ERBB2IP, NTRK2, FSHB, AKAP4 | |
| GOTERM_BP_FAT | GO:0019722~calcium-mediated signaling | 6 | 0.574162679 | 0.045969671 | PLCE1, MCTP2, IL8, ALMS1, NFKBIL1, MCTP1 | |
| GOTERM_BP_FAT | GO:0006468~protein amino acid phosphorylation | 49 | 4.688995215 | 0.023096414 | BMP10, PASK, RPS6KB2, PINK1, TTN, TGFB1, PSKH2, MAP3K5, AAK1, ROS1, IRAK2, RET, MYO3A, PHKG2, MYLK4, SRPK1, CDKL4, MAST4, HUNK, PLCE1, PROK1, RELN, LRRK2, LRRK1, EIF2AK4, KALRN, MAP3K11, FGFR4, ERBB2, STK10, C5, TRIB3, SGK223, TTBK2, EGF, AATK, OBSCN, FSCB, ATR, OXSR1, GUCY2C, EPHA1, EPHA3, GUCY2D, EPHA5, MAPK12, NTRK2, GRK6, CDC42BPB | |
| GOTERM_BP_FAT | Cell cycle process | GO:0022402~cell cycle process | 49 | 4.688995215 | 0.001052892 | MLH3, TTN, TGFB1, KIF2C, DDX11, INCENP, PIWIL3, C11ORF82, TUBB1, ASPM, KIF11, CGRRF1, SGOL2, CCNF, POLE, PLK1S1, MYH9, NCAPD2, REC8, SASS6, PPM1D, FANCD2, BUB1B, HORMAD2, NUP43, DST, MAP3K11, CEP120, TIPIN, CETN3, KIAA1009, PLAGL1, PRUNE2, PSMF1, MACF1, MNS1, GFI1, ZWILCH, NFATC1, IL8, CENPF, BRCA2, TP73, RGS14, PSMB9, CCNB3, TEX15, MAPK12, APBB1 |
| GOTERM_BP_FAT | GO:0030203~glycosaminoglycan metabolic process | 10 | 0.956937799 | 0.002400935 | HYAL2, GCNT2, SPOCK3, XYLT1, HAS1, GALNT5, HEXB, ITIH5, DSE, GLCE | |
| GOTERM_BP_FAT | GO:0007049~cell cycle | 60 | 5.741626794 | 0.003955172 | STEAP3, MLH3, TTN, TGFB1, KIF2C, DDX11, INCENP, PIWIL3, C11ORF82, TUBB1, ASPM, KIF11, CGRRF1, SGOL2, POLE, CCNF, MCM2, PLK1S1, MYH9, AHR, NCAPD2, PPM1D, REC8, SASS6, EP300, RIF1, FANCD2, PRR5-ARHGAP8, BUB1B, TMPRSS11A, HORMAD2, NUP43, DST, MAP3K11, CEP120, CETN3, TIPIN, KIAA1009, PLAGL1, PSMF1, PRUNE2, MACF1, HJURP, MNS1, GFI1, ZWILCH, NFATC1, CKAP2, IL8, CENPF, BRCA2, ATR, RGS14, TP73, PSMB9, CCNB3, TEX15, ERBB2IP, MAPK12, APBB1 | |
| GOTERM_BP_FAT | GO:0070192~chromosome organization involved in meiosis | 4 | 0.38277512 | 0.029295159 | REC8, TEX15, FANCD2, MLH3 | |
| GOTERM_BP_FAT | GO:0000279~M phase | 29 | 2.775119617 | 0.010713242 | CEP120, TIPIN, CETN3, MLH3, TTN, KIAA1009, KIF2C, DDX11, INCENP, MNS1, PIWIL3, ZWILCH, TUBB1, ASPM, KIF11, SGOL2, CCNF, BRCA2, CENPF, MYH9, RGS14, NCAPD2, REC8, CCNB3, TEX15, FANCD2, BUB1B, HORMAD2, NUP43 | |
| GOTERM_BP_FAT | GO:0007059~chromosome segregation | 10 | 0.956937799 | 0.028905796 | REC8, DDX11, HJURP, SGOL2, INCENP, CENPF, TTN, NUP43, SRPK1, NCAPD2 | |
| GOTERM_BP_FAT | GO:0045005~maintenance of fidelity during DNA-dependent DNA replication | 3 | 0.28708134 | 0.025738663 | TIPIN, BRCA2, WRN | |
| GOTERM_BP_FAT | GO:0022403~cell cycle phase | 35 | 3.349282297 | 0.008864905 | CEP120, CETN3, TIPIN, MLH3, TTN, KIAA1009, PRUNE2, KIF2C, DDX11, INCENP, MNS1, PIWIL3, GFI1, ZWILCH, TUBB1, ASPM, NFATC1, KIF11, SGOL2, CCNF, POLE, CENPF, BRCA2, MYH9, RGS14, NCAPD2, CCNB3, REC8, PPM1D, TEX15, FANCD2, BUB1B, HORMAD2, NUP43, MAP3K11 | |
| GOTERM_BP_FAT | Polysaccharide biosynthetic process | GO:0006024~glycosaminoglycan biosynthetic process | 6 | 0.574162679 | 0.004327438 | GCNT2, XYLT1, HAS1, GALNT5, DSE, GLCE |
| GOTERM_BP_FAT | GO:0005976~polysaccharide metabolic process | 14 | 1.339712919 | 0.006372263 | HYAL2, GCNT2, SPOCK3, PHKG2, GALNT5, HEXB, DSE, GLCE, XYLT1, HAS1, MGAM, GAA, ITIH5, AGL | |
| GOTERM_BP_FAT | GO:0000271~polysaccharide biosynthetic process | 8 | 0.765550239 | 0.009446334 | GCNT2, XYLT1, HAS1, GALNT5, PHKG2, DSE, AGL, GLCE | |
| GOTERM_BP_FAT | GO:0006023~aminoglycan biosynthetic process | 6 | 0.574162679 | 0.006548599 | GCNT2, XYLT1, HAS1, GALNT5, DSE, GLCE | |
| GOTERM_BP_FAT | GO:0006022~aminoglycan metabolic process | 10 | 0.956937799 | 0.007510441 | HYAL2, GCNT2, SPOCK3, XYLT1, HAS1, GALNT5, HEXB, ITIH5, DSE, GLCE | |
| GOTERM_BP_FAT | Tissue morphogenesis | GO:0043062~extracellular structure organization | 18 | 1.722488038 | 0.006517427 | RXFP1, PCDHB5, ERBB2, UTRN, HSPG2, NRD1, NID1, COL5A3, COL5A2, SPINK5, COL4A6, TNFRSF11B, COL19A1, CRISPLD2, ACAN, COL12A1, AGRN, APBB1 |
| GOTERM_BP_FAT | GO:0030198~extracellular matrix organization | 13 | 1.244019139 | 0.009627305 | RXFP1, HSPG2, NID1, COL5A3, COL5A2, SPINK5, COL4A6, TNFRSF11B, COL19A1, CRISPLD2, ACAN, COL12A1, APBB1 | |
| GOTERM_BP_FAT | GO:0032989~cellular component morphogenesis | 32 | 3.062200957 | 0.023088526 | BMP10, PLXNA3, SHROOM2, NRP1, COX10, ERBB2, TTN, TGFB1, MACF1, DYNC2H1, OBSL1, GDF9, ROBO3, NFATC1, OPA1, C2CD3, TBCE, ALMS1, MYH6, MYH9, SLIT2, KRT19, ERBB2IP, LAMA5, PRICKLE2, RELN, CNTN4, MAP7, APBB1, DST, CDC42BPB, KALRN | |
| GOTERM_BP_FAT | GO:0048729~tissue morphogenesis | 17 | 1.626794258 | 0.032413281 | DVL2, BMP10, RET, C2CD3, SMAD4, MYH6, NR4A3, TTN, GLI3, TCF7L1, SLIT2, FZD6, MACF1, FREM2, LAMA5, GAA, KLK14 | |
| GOTERM_BP_FAT | GO:0043954~cellular component maintenance | 8 | 0.765550239 | 2.54E-04 | IQCB1, SHROOM2, CDHR1, PCDH15, CNGB1, ACAD11, USH2A, GPR98 | |
| GOTERM_BP_FAT | GO:0048496~maintenance of organ identity | 4 | 0.38277512 | 5.89E-04 | IQCB1, ACAD11, USH2A, GPR98 | |
| GOTERM_BP_FAT | Maintenance of organ identity | GO:0050953~sensory perception of light stimulus | 20 | 1.913875598 | 0.023223977 | IQCB1, OPA1, MYO3A, BBS9, CDHR1, RP2, RP1L1, ALMS1, PCDH15, CNGB1, CDH3, CDS1, GPR98, GUCY2D, EYA4, EYS, HMCN1, IMPG1, ACAD11, USH2A |
| GOTERM_BP_FAT | GO:0007601~visual perception | 20 | 1.913875598 | 0.023223977 | IQCB1, OPA1, MYO3A, BBS9, CDHR1, RP2, RP1L1, ALMS1, PCDH15, CNGB1, CDH3, CDS1, GPR98, GUCY2D, EYA4, EYS, HMCN1, IMPG1, ACAD11, USH2A | |
| GOTERM_BP_FAT | GO:0045494~photoreceptor cell maintenance | 7 | 0.669856459 | 6.29E-04 | IQCB1, CDHR1, PCDH15, CNGB1, ACAD11, USH2A, GPR98 | |
| GOTERM_BP_FAT | GO:0050954~sensory perception of mechanical stimulus | 11 | 1.052631579 | 0.0491677 | KCNQ4, MYO3A, CHRNA9, MCOLN3, HEXB, TRPA1, GJB3, ALMS1, PCDH15, USH2A, GPR98 | |
| GOTERM_BP_FAT | Muscle cell development, differentiation | GO:0045214~sarcomere organization | 4 | 0.38277512 | 0.023449715 | BMP10, KRT19, MYH6, TTN |
| GOTERM_BP_FAT | GO:0030239~myofibril assembly | 5 | 0.4784689 | 0.023617794 | BMP10, KRT19, OBSL1, MYH6, TTN | |
| GOTERM_BP_FAT | GO:0014706~striated muscle tissue development | 13 | 1.244019139 | 0.025697664 | BMP10, ERBB2, UTRN, HSPG2, NRD1, MYH6, TTN, COL19A1, EP300, GAA, OBSL1, ZFPM2, AGRN | |
| GOTERM_BP_FAT | GO:0031032~actomyosin structure organization | 6 | 0.574162679 | 0.015342389 | BMP10, KRT19, LIMCH1, OBSL1, MYH6, TTN | |
| GOTERM_BP_FAT | GO:0051146~striated muscle cell differentiation | 11 | 1.052631579 | 0.019053324 | BMP10, KRT19, ERBB2, UTRN, CACNA1H, NRD1, OBSL1, AGRN, MYH6, MYH9, TTN | |
| GOTERM_BP_FAT | GO:0055001~muscle cell development | 9 | 0.861244019 | 0.009510235 | BMP10, KRT19, ERBB2, UTRN, NRD1, OBSL1, AGRN, MYH6, TTN | |
| GOTERM_BP_FAT | GO:0042692~muscle cell differentiation | 13 | 1.244019139 | 0.028805 | BMP10, ERBB2, UTRN, NRD1, MYH6, TTN, MYH9, SYNE1, KRT19, MAPK12, OBSL1, CACNA1H, AGRN | |
| GOTERM_BP_FAT | GO:0060537~muscle tissue development | 13 | 1.244019139 | 0.03581975 | BMP10, ERBB2, UTRN, HSPG2, NRD1, MYH6, TTN, COL19A1, EP300, GAA, OBSL1, ZFPM2, AGRN | |
| GOTERM_BP_FAT | GO:0030048~actin filament-based movement | 5 | 0.4784689 | 0.032123528 | MYH2, MYH4, MYH14, MYH6, MYH9 | |
| GOTERM_BP_FAT | GO:0055002~striated muscle cell development | 9 | 0.861244019 | 0.006074436 | BMP10, KRT19, ERBB2, UTRN, NRD1, OBSL1, AGRN, MYH6, TTN | |
| GOTERM_BP_FAT | GO:0007517~muscle organ development | 22 | 2.105263158 | 0.00464554 | BMP10, AEBP1, ERBB2, UTRN, HSPG2, ITGA11, CENPF, NRD1, MYH6, TTN, EP300, COL19A1, MAPK12, NEB, LAMA5, GAA, CACNA1H, OBSL1, ZFPM2, AGRN, UNC45B, UNC45A | |
| GOTERM_BP_FAT | GO:0048747~muscle fiber development | 6 | 0.574162679 | 0.037333701 | ERBB2, UTRN, NRD1, AGRN, MYH6, TTN | |
| GOTERM_BP_FAT | GO:0007507~heart development | 19 | 1.818181818 | 0.040567691 | DVL2, BMP10, NRP1, C2CD3, ERBB2, HSPG2, OXTR, MYH6, TTN, GLI3, PLCE1, EP300, SALL4, GAA, OBSL1, ZFPM2, ADAM19, NFATC3, NFATC1 | |
| GOTERM_BP_FAT | Negative regulation | GO:0050860~negative regulation of T cell receptor signaling pathway | 3 | 0.28708134 | 0.025738663 | ELF1, CBLB, UBASH3A |
| GOTERM_BP_FAT | GO:0050858~negative regulation of antigen receptor-mediated signaling pathway | 3 | 0.28708134 | 0.025738663 | ELF1, CBLB, UBASH3A | |
| GOTERM_BP_FAT | GO:0010596~negative regulation of endothelial cell migration | 4 | 0.38277512 | 0.035834358 | BMP10, AGTR2, DLL4, TGFB1 | |
| GOTERM_BP_FAT | GO:0045792~negative regulation of cell size | 11 | 1.052631579 | 0.039161459 | RTN4, BMP10, PLXNA3, AGTR2, NRP1, CGRRF1, SMAD4, GDF9, APBB1, TP73, TGFB1 | |
| GOTERM_BP_FAT | GO:0030308~negative regulation of cell growth | 11 | 1.052631579 | 0.025196073 | RTN4, BMP10, PLXNA3, AGTR2, NRP1, CGRRF1, SMAD4, GDF9, APBB1, TP73, TGFB1 | |
| GOTERM_BP_FAT | Cell death | GO:0008219~cell death | 51 | 4.880382775 | 0.036510953 | STEAP3, RTN4, TSPO, FASTKD1, TGFB1, MAGED1, TNFRSF11B, MAP3K5, TIAM1, CLUL1, C11ORF82, CASP1, API5, MAGI3, OPA1, GZMA, PTPRH, SCN2A, CECR2, ARHGEF17, AHR, EP300, RASGRF2, ZFYVE27, ZFYVE26, SH3KBP1, BUB1B, KALRN, MAP3K11, C5, TRIB3, PRUNE2, PLEKHG2, ATN1, TTBK2, TRAF5, HIP1, AATK, CKAP2, OBSCN, CARD8, VAV3, ALMS1, CIDEA, FIG4, TP73, NFKBIL1, ATXN3, SYNE1, PARP4, APBB1 |
| GOTERM_BP_FAT | GO:0016265~death | 51 | 4.880382775 | 0.039214579 | STEAP3, RTN4, TSPO, FASTKD1, TGFB1, MAGED1, TNFRSF11B, MAP3K5, TIAM1, CLUL1, C11ORF82, CASP1, API5, MAGI3, OPA1, GZMA, PTPRH, SCN2A, CECR2, ARHGEF17, AHR, EP300, RASGRF2, ZFYVE27, ZFYVE26, SH3KBP1, BUB1B, KALRN, MAP3K11, C5, TRIB3, PRUNE2, PLEKHG2, ATN1, TTBK2, TRAF5, HIP1, AATK, CKAP2, OBSCN, CARD8, VAV3, ALMS1, CIDEA, FIG4, TP73, NFKBIL1, ATXN3, SYNE1, PARP4, APBB1 | |
| GOTERM_BP_FAT | Other | GO:0009566~fertilization | 10 | 0.956937799 | 0.025036283 | ACR, PLCZ1, APOB, TEX15, ZP2, CD46, HEXB, UBXN8, KLK14, AKAP4 |
| GOTERM_BP_FAT | GO:0007129~synapsis | 4 | 0.38277512 | 0.029295159 | REC8, TEX15, FANCD2, MLH3 | |
| KEGG_PATHWAY | KEGG pathway | hsa02010:ABC transporters | 10 | 0.956937799 | 4.87E-04 | ABCA10, ABCG5, TAP1, ABCC4, ABCC10, ABCC1, ABCB5, ABCA6, ABCA13, ABCB4 |
| KEGG_PATHWAY | hsa04512:ECM-receptor interaction | 14 | 1.339712919 | 5.35E-04 | COL4A1, HSPG2, ITGA11, COL5A3, COL5A2, COL4A6, HMMR, VWF, LAMB3, LAMA3, LAMA5, COL6A1, RELN, AGRN | |
| KEGG_PATHWAY | hsa04142:Lysosome | 13 | 1.244019139 | 0.024611161 | ARSB, AP1B1, HEXB, ASAH1, SLC11A1, NPC1, NAGPA, LAPTM5, IGF2R, GAA, CTSB, ATP6V0D2, GGA3 | |
| KEGG_PATHWAY | hsa04610:Complement and coagulation cascades | 9 | 0.861244019 | 0.032655465 | F11, VWF, CR2, MASP1, FGA, C3, CD46, C5, BDKRB2 | |
| KEGG_PATHWAY | hsa00630:Glyoxylate and dicarboxylate metabolism | 4 | 0.38277512 | 0.044700865 | MTHFD1, ACO1, HAO2, GRHPR |
Abbreviations: ECM, extracellular matrix.
Figure 2.
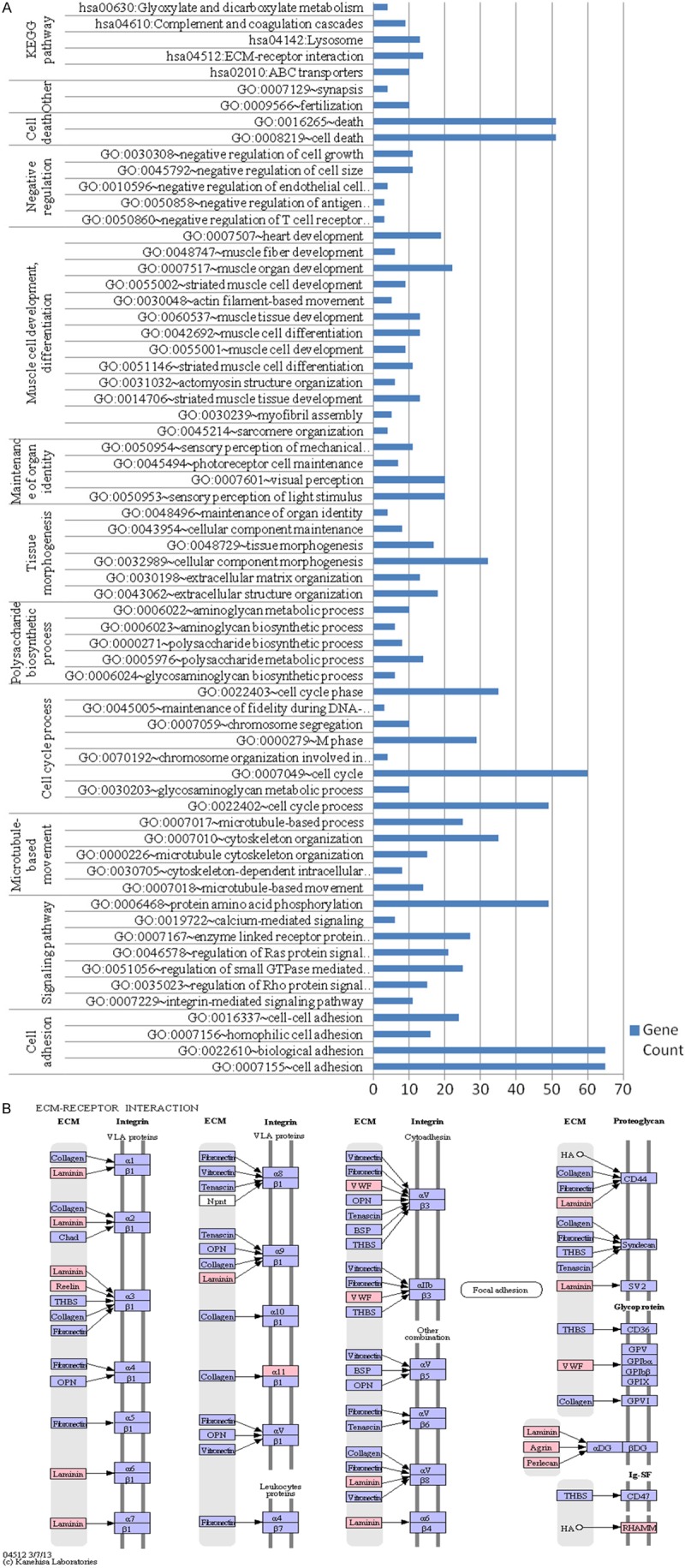
Functional enrichment analysis of the 211 missense-mutated genes detected by exome sequencing in PRCC. A. The related biological process categories of the 211 missense-mutated genes in CRCC. B. The extracellular matrix (ECM)-receptor interaction pathway. red genes, mutated genes in PRCC. Pathway information was generated using the Kyoto Encyclopedia of Genes and Genomes database.
The missense mutation status of 19 genes was significantly different (P < 0.05) between the type 1 PRCC C and type 2 PRCC groups (Table 5). Alterations in EEF1D, RFNG, GPR142, and RAB37 genes were located in different chromosomal regions in the type 1 PRCC C and type 2 PRCC groups.
Table 5.
The 19 differentially missense mutated genes in type 1PRCC C vs. type 2 PRCC (P < 0.05)**
| SNP_name | Chr | Alleles | Mutation (s) | Gene |
|---|---|---|---|---|
| exm330459 | 3p12.3 | [C/G] | Missense_H75D | CNTN3 |
| exm318874 | 3p21.2 | [A/G] | Missense_R425C, Missense_R426C | VPRBP |
| exm506256 | 5q35.2 | [A/G] | Missense_A328T, Missense_A328T, Missense_A328T | FGFR4 |
| exm611166 | 7p15.2 | [C/G] | Missense_R132S | HOXA11 |
| exm693941 | 8p12 | [A/G] | Missense_T2181I | TEX15 |
| exm727114 | 8q24.3 | [A/C] | Missense_L361R, Missense_L361R | EEF1D |
| exm919007 | 11q12.3 | [G/C] | Missense_A866P | INTS5 |
| exm940191 | 11q13.4 | [A/G] | Missense_R142Q | DNAJB13 |
| exm976848 | 12p13.3 | [T/C] | Missense_R606Q | VWF |
| exm1185487 | 15q24-q25 | [A/G] | Missense_D1086N, Missense_D1086N | AKAP13 |
| exm1368709 | 17q25 | [A/C] | Missense_H288Q | RFNG |
| exm1277466 | 17p13.3 | [T/C] | Missense_P285S | OR1A1 |
| exm1351674 | 17q25.1 | [T/C] | Missense_T407M | GPR142 |
| exm1352075 | 17q25.1 | [T/G] | Missense_T282K | RAB37 |
| exm1379777 | 18q11.2 | [A/G] | Missense_A152T | TAF4B |
| exm1395964 | 19p13.3 | [T/C] | Missense_A314V, Missense_A227V | MADCAM1 |
| exm1529410 | 20p11.21 | [T/C] | Missense_P297S | GZF1 |
| exm1663015 | Xq28 | [T/C] | Missense_V377A | PNMA3 |
Type 1 PRCC C vs. type 2 PRCC; Fisher’s exact test.
Abbreviations: PRCC, Papillary renal cell carcinoma; Chr., chromosome; SNP, single nucleotide polymorphism.
Discussion
PRCC is the second most prevalent renal tumor after renal clear cell carcinoma [1]. PRCC can be divided into two types based on the histomorphological features. The onset age and sex of PRCC patients are similar to ccRCC patients, with a peak incidence in 50-70-year-old men [1,11]. Herein, the average age of the patients was 53.9 years (range, 26-74 years). Compared with type 1, the mean age of type 2 PRCC patients was approximately 14 years lower (57.4 vs. 61.5 years), which is consistent with the results of previous studies [12,13].
Pathologically, type 2 tumors showed a higher Fuhrman grade (P = 0.049) and lymphovascular invasion (P = 0.049) than type 1, which have both been identified as prognostic factors [14,15], suggesting poorer outcomes in type 2 PRCC patients. While some studies have reported no clear correlation between PRCC type and prognosis [16,17], most have shown that type 1 PRCC has a better prognosis compared to type 2 [11-13,18]. Moreover, compared with the overall survival rates of patients with type 1 PRCC, those of type 2 PRCC were lower in this study, suggesting that tumor classification is indeed helpful for evaluating the prognosis of PRCC patients.
Immunohistochemically, all 13 tumors showed strong positivity for AMACR, while CK7 and Top IIα were overexpressed in types 1 and 2 PRCC, respectively. This is consistent with previous reports [1,19-21], suggesting that AMACR, CK7, and Top IIα are useful for the classification, diagnosis, and differential diagnosis of PRCC. Importantly, increased Top IIα expression correlates to poorer prognosis of various tumors, such as breast and colon cancer [22,23], and some researchers found that Top IIα expression is increased in type 2 PRCC with higher Fuhrman nuclear grade, and that the level of Top IIα positively correlates with tumor invasion [24]. Herein, type 1 PRCC did not express Top IIα, whereas 57.1% of type 2 PRCC cases did (4/7), indicating that Top IIα not only contributes to the differential diagnosis, classification, and prognosis of PRCC, but may also play a role in its development.
In order to further detect gene mutations, we analyzed the exon of 13 PRCC and 18 normal kidney tissues by whole-genome exon sequencing. In the cluster analysis, we identified 10 enriched clusters (Table 4), with the frequency of gene mutations related to the cell division cycle being the highest (Figure 2A). Cell division is an important process, and problems during the processing can result in abnormal cell division, proliferation, differentiation, and senescence. Numerous growth factors, cytokines, hormones, and cancer gene products regulate metabolism by influencing the cell division cycle. Meanwhile, the expression of many genes is restricted by the cell division cycle. Thus, our results suggest that these genes may play important roles in the occurrence and development of PRCC.
In the cell division cycle cluster, many interesting genes, such as MAP3K11 and KIF11, were identified. The protein encoded by MAP3K11 may activate MAPK8/JNK kinase, which regulates the JNK signal pathway and activates NF-kappa B signaling pathway, mediated by GTPases and CDC42, which in turn regulates cell proliferation and apoptosis [25,26]. Recently, MAP3K11 has been shown to play a role in the development of prostate, breast, and gastric cancers through interfering with cell proliferation and apoptosis [27-29]. KIF11 encodes a kinesin spindle protein, a member of the kinesin superfamily of microtubule-based motors, and plays a critical role in mitosis through mediation of centrosome separation and bipolar spindle assembly and maintenance. Reduced KIF11 expression leads to cell cycle arrest at mitosis and formation of monoastral microtubule arrays, and, ultimately, to tumor cell death [30-32]. Sun et al. [33] reported that KIF11 overexpression correlated with nuclear grade (P = 0.019), stage (P = 0.007), and tumor size (P = 0.033) in RCC, and as type 2 PRCC shows higher nuclear grade and stage and worse prognosis than type 1, it can be speculated that it is associated with MAP3K11 and KIF11 mutations; however, further studies are needed to confirm this hypothesis.
The pathway enrichment analysis revealed 5 related pathways (Table 4), with the “ABC transporter” pathway being the most significant pathway in PRCC. The ABC transporters form one of the largest known protein families, and couple ATP hydrolysis to active transport of a wide variety of substrates such as lipids, sterols, proteins, and drugs. Numerous studies have shown that this pathway plays an important role in the development of multi-drug resistant tumors [34-36]. These proteins can actively transport drugs from the intracellular to extracellular compartments, thereby reducing the intracellular concentration of drugs. Zhao et al. [36] showed that ABCC4 was highly expressed in lung cancer, and that reduced ABCC4 expression could inhibit tumor growth and proliferation. Walsh et al. [37] showed that ABCB1 and ABCC1 up regulation resulted in the development of multi-drug resistant RCC, and Hour et al. [38] reported that ABCD1 down regulation may be involved in renal tumorigenesis. Therefore, we inferred that mutations in the ABC pathways may reduce the effectiveness of chemotherapy drugs and promote the growth and proliferation of PRCC cells, and that inhibition of the ABC transporters may increase the efficacy of chemotherapy and slow down the development of PRCC.
Additionally, in the 5 related pathways, “ECM-receptor interaction” mutations commonly occurred (Figure 2B), with the mutation frequency of the collagen family genes being the highest. COL4A1 encodes the major type IV alpha collagen chain of basement membranes, which plays an essential role in tumorigenesis, growth, and metastasis. Delektorskaya et al. [39] suggested that type IV collagen shows different degrees of loss in colorectal cancer, which significantly correlated with the risk of metastasis. Others have found that type IV collagen promotes tumor cell migration and invasion in pancreatic cancer, and that the level of serum type IV collagen in these patients positively correlated with the risk of recurrence [40,41]. Moreover, RCC cells can also produce type IV collagen as a means to promote tumor invasion [42-44], indicating that the frequent mutations of the collagen genes may be one of factors responsible for the development of PRCC, and that evaluating the collagen levels of PRCC patient may be useful for assessing the tumor biological behavior and prognosis.
There have reports that type 1 and 2 PRCC show more copy number changes at 17q and 9p [45-48]. Furthermore, copy number changes at 17q were more common in TNM stage 1-2 PRCC and correlated with lower stage, less lymphatic metastases, and increased survival, whereas changes at 9p conversely correlated with higher stage (TNM stage 3-4) and nuclear grade, more lymphatic metastasis, and decreased survival [45-47]. Meanwhile, amplification of chromosome 17 is another characteristic of PRCC [45,49,50], and changes at 17q and 9p can aid the differential diagnosis, as well as predict the prognosis in different subtypes, suggesting that genes on these chromosomes may be related to the development of type 1 or 2 PRCC. The results from exon chip analyses are consistent with previous reports in the field, with some gene exon mutations being found in specific altered chromosomal regions. For example, ERBB2 locate on 17q12-20. ERBB2 encodes a member of the tyrosine kinase family. It is over expressed or amplified in several tumors, including breast, ovarian, and digestive tract tumors, and closely correlates with tumor occurrence, development, and prognosis [51]. Conversely, the over expression and amplification of ERBB2 is reportedly uncommon in RCC [52,53]. However, Duzcan et al. [54] found that the levels of Top IIα and ERBB2 were correlated, and that they were co-amplified. Herein, Top IIα was found to be over expressed in type 2 PRCC, and located on the common aberration chromosome 3p24; ERBB2 is located at 17q12-20, which showed amplification, and exon chip detection moreover revealed ERBB2 mutations. This suggests that Top IIα and ERBB2 may jointly participate in the occurrence and development of PRCC, and that exon chip analyses may facilitate the discovery of mutated genes in PRCC.
Using exome sequencing, we here found that the EEF1D, RFNG, GPR142, and RAB37 genes were located in different chromosomal regions in type 1 and 2 PRCC. RAB37, which is located at chromosome 17q25.1, more often showed gains in type 1 PRCC. Dobashi et al. [55] found that RAB37 was upregulated in RCC cells, and knockdown of RAB37 expression by specific siRNA caused significant reductions in cancer cell growth. Furthermore, Wu [56] also found that promoter/exon 1 methylation lead to down-regulation of hRAB37 in metastatic lung cancer, and that it may serve as a predictive biomarker of lung cancer progression. EEF1D, which is located at chromosome 8q24.3 and was more commonly mutated in type 2 PRCC, is also overexpressed in medulloblastoma [57] and right-sided colon cancer [58], and correlates with the invasive status of adriamycin-resistant variants of DLKP, a squamous lung cancer cell line [59]. Accordingly, we speculate that the mutations of RAB37 and EEF1D may play different roles in the development of type 1 and 2 PRCC.
In conclusion, our study shows that multiple gene mutations are present in PRCC. These gene mutations may provide clues regarding PRCC tumorigenesis and serve as a basis for future developments of targeted therapies against type 1 and 2 PRCC.
Acknowledgements
Supported by grants from the National Natural Science Foundation of China (NSFC, No. 81060383). We would like to thank Editage http://www.editage.cn/ for English language editing.
Disclosure of conflict of interest
None.
References
- 1.John N, Eble GS, Jonathan I, Epstein Isabell A, Sesterhenn . World health organization classification of tumors: pathology and genetics of tumors of the urinary system and male genital organs. Lyon: IARC Press; 2004. [Google Scholar]
- 2.Delahunt B, Eble JN, McCredie MR, Bethwaite PB, Stewart JH, Bilous AM. Morphologic typing of papillary renal cell carcinoma: comparison of growth kinetics and patient survival in 66 cases. Hum Pathol. 2001;32:590–595. doi: 10.1053/hupa.2001.24984. [DOI] [PubMed] [Google Scholar]
- 3.Steffens S, Janssen M, Roos FC, Becker F, Schumacher S, Seidel C, Wegener G, Thuroff JW, Hofmann R, Stockle M, Siemer S, Schrader M, Hartmann A, Kuczyk MA, Junker K, Schrader AJ. Incidence and long-term prognosis of papillary compared to clear cell renal cell carcinoma--a multicentre study. Eur J Cancer. 2012;48:2347–2352. doi: 10.1016/j.ejca.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J Surg Pathol. 2003;27:612–624. doi: 10.1097/00000478-200305000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Ficarra V, Martignoni G, Galfano A, Novara G, Gobbo S, Brunelli M, Pea M, Zattoni F, Artibani W. Prognostic role of the histologic subtypes of renal cell carcinoma after slide revision. Eur Urol. 2006;50:786–793. doi: 10.1016/j.eururo.2006.04.009. discussion 793-784. [DOI] [PubMed] [Google Scholar]
- 6.Gordon MS, Hussey M, Nagle RB, Lara PN Jr, Mack PC, Dutcher J, Samlowski W, Clark JI, Quinn DI, Pan CX, Crawford D. Phase II study of erlotinib in patients with locally advanced or metastatic papillary histology renal cell cancer: SWOG S0317. J. Clin. Oncol. 2009;27:5788–5793. doi: 10.1200/JCO.2008.18.8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patard JJ, Leray E, Rioux-Leclercq N, Cindolo L, Ficarra V, Zisman A, De La Taille A, Tostain J, Artibani W, Abbou CC, Lobel B, Guille F, Chopin DK, Mulders PF, Wood CG, Swanson DA, Figlin RA, Belldegrun AS, Pantuck AJ. Prognostic value of histologic subtypes in renal cell carcinoma: a multicenter experience. J. Clin. Oncol. 2005;23:2763–2771. doi: 10.1200/JCO.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 8.Ridge CA, Pua BB, Madoff DC. Epidemiology and staging of renal cell carcinoma. Semin Intervent Radiol. 2014;31:3–8. doi: 10.1055/s-0033-1363837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, Scherer SW, Zhuang Z, Lubensky I, Dean M, Allikmets R, Chidambaram A, Bergerheim UR, Feltis JT, Casadevall C, Zamarron A, Bernues M, Richard S, Lips CJ, Walther MM, Tsui LC, Geil L, Orcutt ML, Stackhouse T, Lipan J, Slife L, Brauch H, Decker J, Niehans G, Hughson MD, Moch H, Storkel S, Lerman MI, Linehan WM, Zbar B. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 10.Farber LJ, Furge K, Teh BT. Renal cell carcinoma deep sequencing: recent developments. Curr Oncol Rep. 2012;14:240–248. doi: 10.1007/s11912-012-0230-3. [DOI] [PubMed] [Google Scholar]
- 11.Kosaka T, Mikami S, Miyajima A, Kikuchi E, Nakagawa K, Ohigashi T, Nakashima J, Oya M. Papillary renal cell carcinoma: clinicopathological characteristics in 40 patients. Clin Exp Nephrol. 2008;12:195–199. doi: 10.1007/s10157-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 12.Antonelli A, Tardanico R, Balzarini P, Arrighi N, Perucchini L, Zanotelli T, Cozzoli A, Zani D, Cunico SC, Simeone C. Cytogenetic features, clinical significance and prognostic impact of type 1 and type 2 papillary renal cell carcinoma. Cancer Genet Cytogenet. 2010;199:128–133. doi: 10.1016/j.cancergencyto.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Yamanaka K, Miyake H, Hara I, Inoue TA, Hanioka K, Fujisawa M. Papillary renal cell carcinoma: a clinicopathological study of 35 cases. Int J Urol. 2006;13:1049–1052. doi: 10.1111/j.1442-2042.2006.01500.x. [DOI] [PubMed] [Google Scholar]
- 14.Margulis V, Tamboli P, Matin SF, Swanson DA, Wood CG. Analysis of clinicopathologic predictors of oncologic outcome provides insight into the natural history of surgically managed papillary renal cell carcinoma. Cancer. 2008;112:1480–1488. doi: 10.1002/cncr.23322. [DOI] [PubMed] [Google Scholar]
- 15.Zucchi A, Novara G, Costantini E, Antonelli A, Carini M, Carmignani G, Cosciani Cunico S, Fontana D, Longo N, Martignoni G, Minervini A, Mirone V, Porena M, Roscigno M, Schiavina R, Simeone C, Simonato A, Siracusano S, Terrone C, Ficarra V. Prognostic factors in a large multi-institutional series of papillary renal cell carcinoma. BJU Int. 2012;109:1140–1146. doi: 10.1111/j.1464-410X.2011.10517.x. [DOI] [PubMed] [Google Scholar]
- 16.Mejean A, Hopirtean V, Bazin JP, Larousserie F, Benoit H, Chretien Y, Thiounn N, Dufour B. Prognostic factors for the survival of patients with papillary renal cell carcinoma: meaning of histological typing and multifocality. J Urol. 2003;170:764–767. doi: 10.1097/01.ju.0000081122.57148.ec. [DOI] [PubMed] [Google Scholar]
- 17.Allory Y, Ouazana D, Boucher E, Thiounn N, Vieillefond A. Papillary renal cell carcinoma. Prognostic value of morphological subtypes in a clinicopathologic study of 43 cases. Virchows Arch. 2003;442:336–342. doi: 10.1007/s00428-003-0787-1. [DOI] [PubMed] [Google Scholar]
- 18.Waldert M, Haitel A, Marberger M, Katzenbeisser D, Ozsoy M, Stadler E, Remzi M. Comparison of type I and II papillary renal cell carcinoma (RCC) and clear cell RCC. BJU Int. 2008;102:1381–1384. doi: 10.1111/j.1464-410X.2008.07999.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Williamson SR, Wang M, Davidson DD, Zhang S, Baldridge LA, Du X, Cheng L. Molecular subtyping of metastatic renal cell carcinoma: implications for targeted therapy. Mol Cancer. 2014;13:39. doi: 10.1186/1476-4598-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Ahmadie HA, Alden D, Fine SW, Gopalan A, Touijer KA, Russo P, Reuter VE, Tickoo SK. Role of immunohistochemistry in the evaluation of needle core biopsies in adult renal cortical tumors: an ex vivo study. Am J Surg Pathol. 2011;35:949–961. doi: 10.1097/PAS.0b013e31821e25cd. [DOI] [PubMed] [Google Scholar]
- 21.Williamson SR, Halat S, Eble JN, Grignon DJ, Lopez-Beltran A, Montironi R, Tan PH, Wang M, Zhang S, Maclennan GT, Baldridge LA, Cheng L. Multilocular cystic renal cell carcinoma: similarities and differences in immunoprofile compared with clear cell renal cell carcinoma. Am J Surg Pathol. 2012;36:1425–1433. doi: 10.1097/PAS.0b013e31825b37f0. [DOI] [PubMed] [Google Scholar]
- 22.Panousis D, Patsouris E, Lagoudianakis E, Pappas A, Kyriakidou V, Voulgaris Z, Xepapadakis G, Manouras A, Athanassiadou AM, Athanassiadou P. The value of TOP2A, EZH2 and paxillin expression as markers of aggressive breast cancer: relationship with other prognostic factors. Eur J Gynaecol Oncol. 2011;32:156–159. [PubMed] [Google Scholar]
- 23.Gao XH, Yu ZQ, Zhang C, Bai CG, Zheng JM, Fu CG. DNA topoisomerase II alpha: a favorable prognostic factor in colorectal caner. Int J Colorectal Dis. 2012;27:429–435. doi: 10.1007/s00384-011-1346-x. [DOI] [PubMed] [Google Scholar]
- 24.Dekel Y, Frede T, Kugel V, Neumann G, Rassweiler J, Koren R. Human DNA topoisomerase II-alpha expression in laparoscopically treated renal cell carcinoma. Oncol Rep. 2005;14:271–274. [PubMed] [Google Scholar]
- 25.Ding S, Xing N, Lu J, Zhang H, Nishizawa K, Liu S, Yuan X, Qin Y, Liu Y, Ogawa O, Nishiyama H. Overexpression of Eg5 predicts unfavorable prognosis in non-muscle invasive bladder urothelial carcinoma. Int J Urol. 2011;18:432–438. doi: 10.1111/j.1442-2042.2011.02751.x. [DOI] [PubMed] [Google Scholar]
- 26.Liou GY, Zhang H, Miller EM, Seibold SA, Chen W, Gallo KA. Induced, selective proteolysis of MLK3 negatively regulates MLK3/JNK signalling. Biochem J. 2010;427:435–443. doi: 10.1042/BJ20091077. [DOI] [PubMed] [Google Scholar]
- 27.Whitworth H, Bhadel S, Ivey M, Conaway M, Spencer A, Hernan R, Holemon H, Gioeli D. Identification of kinases regulating prostate cancer cell growth using an RNAi phenotypic screen. PLoS One. 2012;7:e38950. doi: 10.1371/journal.pone.0038950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Miller EM, Gallo KA. MLK3 is critical for breast cancer cell migration and promotes a malignant phenotype in mammary epithelial cells. Oncogene. 2010;29:4399–4411. doi: 10.1038/onc.2010.198. [DOI] [PubMed] [Google Scholar]
- 29.Mishra P, Senthivinayagam S, Rangasamy V, Sondarva G, Rana B. Mixed lineage kinase-3/JNK1 axis promotes migration of human gastric cancer cells following gastrin stimulation. Mol Endocrinol. 2010;24:598–607. doi: 10.1210/me.2009-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martens-de Kemp SR, Nagel R, Stigter-van Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ, Brakenhoff RH. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin Cancer Res. 2013;19:1994–2003. doi: 10.1158/1078-0432.CCR-12-2539. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y, Orth JD, Xie T, Mitchison TJ. Rapid induction of apoptosis during Kinesin-5 inhibitor-induced mitotic arrest in HL60 cells. Cancer Lett. 2011;310:15–24. doi: 10.1016/j.canlet.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marra E, Palombo F, Ciliberto G, Aurisicchio L. Kinesin spindle protein SiRNA slows tumor progression. J Cell Physiol. 2013;228:58–64. doi: 10.1002/jcp.24103. [DOI] [PubMed] [Google Scholar]
- 33.Sun D, Lu J, Ding K, Bi D, Niu Z, Cao Q, Zhang J, Ding S. The expression of Eg5 predicts a poor outcome for patients with renal cell carcinoma. Med Oncol. 2013;30:476. doi: 10.1007/s12032-013-0476-0. [DOI] [PubMed] [Google Scholar]
- 34.Kovalev AA, Tsvetaeva DA, Grudinskaja TV. Role of ABC-cassette transporters (MDR1, MRP1, BCRP) in the development of primary and acquired multiple drug resistance in patients with early and metastatic breast cancer. Exp Oncol. 2013;35:287–290. [PubMed] [Google Scholar]
- 35.Wang F, Wang XK, Shi CJ, Zhang H, Hu YP, Chen YF, Fu LW. Nilotinib enhances the efficacy of conventional chemotherapeutic drugs in CD34 (+) CD38 (-) stem cells and ABC transporter overexpressing leukemia cells. Molecules. 2014;19:3356–3375. doi: 10.3390/molecules19033356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Guo Y, Yue W, Zhang L, Gu M, Wang Y. ABCC4 is required for cell proliferation and tumorigenesis in non-small cell lung cancer. Onco Targets Ther. 2014;7:343–351. doi: 10.2147/OTT.S56029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh N, Larkin A, Kennedy S, Connolly L, Ballot J, Ooi W, Gullo G, Crown J, Clynes M, O’Driscoll L. Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol. 2009;9:6. doi: 10.1186/1471-2490-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hour TC, Kuo YZ, Liu GY, Kang WY, Huang CY, Tsai YC, Wu WJ, Huang SP, Pu YS. Downregulation of ABCD1 in human renal cell carcinoma. Int J Biol Markers. 2009;24:171–178. doi: 10.1177/172460080902400307. [DOI] [PubMed] [Google Scholar]
- 39.Delektorskaya VV, Golovkov DA, Kushlinskii NE. Clinical significance of levels of molecular biological markers in zones of invasive front-line of colorectal cancer. Bull Exp Biol Med. 2008;146:616–619. doi: 10.1007/s10517-009-0343-3. [DOI] [PubMed] [Google Scholar]
- 40.Ohlund D, Lundin C, Ardnor B, Oman M, Naredi P, Sund M. Type IV collagen is a tumour stroma-derived biomarker for pancreas cancer. Br J Cancer. 2009;101:91–97. doi: 10.1038/sj.bjc.6605107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryschich E, Khamidjanov A, Kerkadze V, Buchler MW, Zoller M, Schmidt J. Promotion of tumor cell migration by extracellular matrix proteins in human pancreatic cancer. Pancreas. 2009;38:804–810. doi: 10.1097/MPA.0b013e3181b9dfda. [DOI] [PubMed] [Google Scholar]
- 42.Kato Y, Sakai N, Baba M, Kaneko S, Kondo K, Kubota Y, Yao M, Shuin T, Saito S, Koshika S, Kawase T, Miyagi Y, Aoki I, Nagashima Y. Stimulation of motility of human renal cell carcinoma by SPARC/Osteonectin/BM-40 associated with type IV collagen. Invasion Metastasis. 1998;18:105–114. doi: 10.1159/000024503. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama Y, Naito S, Ryuto M, Hata Y, Ono M, Sueishi K, Komiyama S, Itoh H, Kuwano M. An in vitro invasion model for human renal cell carcinoma cell lines mimicking their metastatic abilities. Clin Exp Metastasis. 1996;14:466–474. doi: 10.1007/BF00128963. [DOI] [PubMed] [Google Scholar]
- 44.Lohi J, Korhonen M, Leivo I, Kangas L, Tani T, Kalluri R, Miner JH, Lehto VP, Virtanen I. Expression of type IV collagen alpha1(IV)-alpha6(IV) polypeptides in normal and developing human kidney and in renal cell carcinomas and oncocytomas. Int J Cancer. 1997;72:43–49. doi: 10.1002/(sici)1097-0215(19970703)72:1<43::aid-ijc6>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 45.Sanders ME, Mick R, Tomaszewski JE, Barr FG. Unique patterns of allelic imbalance distinguish type 1 from type 2 sporadic papillary renal cell carcinoma. Am J Pathol. 2002;161:997–1005. doi: 10.1016/S0002-9440(10)64260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuda D, Khoo SK, Massie A, Iwamura M, Chen J, Petillo D, Wondergem B, Avallone M, Kloostra SJ, Tan MH, Koeman J, Zhang Z, Kahnoski RJ French Kidney Cancer Study Group. Baba S, Teh BT. Identification of copy number alterations and its association with pathological features in clear cell and papillary RCC. Cancer Lett. 2008;272:260–267. doi: 10.1016/j.canlet.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Klatte T, Pantuck AJ, Said JW, Seligson DB, Rao NP, LaRochelle JC, Shuch B, Zisman A, Kabbinavar FF, Belldegrun AS. Cytogenetic and molecular tumor profiling for type 1 and type 2 papillary renal cell carcinoma. Clin Cancer Res. 2009;15:1162–1169. doi: 10.1158/1078-0432.CCR-08-1229. [DOI] [PubMed] [Google Scholar]
- 48.Yang XJ, Tan MH, Kim HL, Ditlev JA, Betten MW, Png CE, Kort EJ, Futami K, Furge KA, Takahashi M, Kanayama HO, Tan PH, Teh BS, Luan C, Wang K, Pins M, Tretiakova M, Anema J, Kahnoski R, Nicol T, Stadler W, Vogelzang NG, Amato R, Seligson D, Figlin R, Belldegrun A, Rogers CG, Teh BT. A molecular classification of papillary renal cell carcinoma. Cancer Res. 2005;65:5628–5637. doi: 10.1158/0008-5472.CAN-05-0533. [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Beltran A, Montironi R, Egevad L, Caballero-Vargas MT, Scarpelli M, Kirkali Z, Cheng L. Genetic profiles in renal tumors. Int J Urol. 2010;17:6–19. doi: 10.1111/j.1442-2042.2009.02395.x. [DOI] [PubMed] [Google Scholar]
- 50.Cheng L, Zhang S, MacLennan GT, Lopez-Beltran A, Montironi R. Molecular and cytogenetic insights into the pathogenesis, classification, differential diagnosis, and prognosis of renal epithelial neoplasms. Hum Pathol. 2009;40:10–29. doi: 10.1016/j.humpath.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Menard S, Casalini P, Campiglio M, Pupa S, Agresti R, Tagliabue E. HER2 overexpression in various tumor types, focussing on its relationship to the development of invasive breast cancer. Ann Oncol. 2001;12(Suppl 1):S15–19. doi: 10.1093/annonc/12.suppl_1.s15. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Liu C, Han J, Zhen L, Zhang T, He X, Xu E, Li M. HER2 expression in renal cell carcinoma is rare and negatively correlated with that in normal renal tissue. Oncol Lett. 2012;4:194–198. doi: 10.3892/ol.2012.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Latif Z, Watters AD, Bartlett JM, Underwood MA, Aitchison M. Gene amplification and overexpression of HER2 in renal cell carcinoma. BJU Int. 2002;89:5–9. [PubMed] [Google Scholar]
- 54.Duzcan F, Duzcan SE, Sen S, Yorukoglu K, Caner V, Sen Turk N, Cetin GO, Kelten C, Tuna B, Sarsik B, Tepeli E. Expression and amplification of Topoisomerase-2alpha in type 1 and type 2 papillary renal cell carcinomas and its correlation with HER2/neu amplification. Pathol Oncol Res. 2011;17:697–703. doi: 10.1007/s12253-011-9372-0. [DOI] [PubMed] [Google Scholar]
- 55.Dobashi S, Katagiri T, Hirota E, Ashida S, Daigo Y, Shuin T, Fujioka T, Miki T, Nakamura Y. Involvement of TMEM22 overexpression in the growth of renal cell carcinoma cells. Oncol Rep. 2009;21:305–312. [PubMed] [Google Scholar]
- 56.Wu CY, Tseng RC, Hsu HS, Wang YC, Hsu MT. Frequent down-regulation of hRAB37 in metastatic tumor by genetic and epigenetic mechanisms in lung cancer. Lung Cancer. 2009;63:360–367. doi: 10.1016/j.lungcan.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 57.De Bortoli M, Castellino RC, Lu XY, Deyo J, Sturla LM, Adesina AM, Perlaky L, Pomeroy SL, Lau CC, Man TK, Rao PH, Kim JY. Medulloblastoma outcome is adversely associated with overexpression of EEF1D, RPL30, and RPS20 on the long arm of chromosome 8. BMC Cancer. 2006;6:223. doi: 10.1186/1471-2407-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen H, Huang J, Pei H, Zeng S, Tao Y, Shen L, Zeng L, Zhu H. Comparative proteomic study for profiling differentially expressed proteins between Chinese left- and right-sided colon cancers. Cancer Sci. 2013;104:135–141. doi: 10.1111/cas.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keenan J, Murphy L, Henry M, Meleady P, Clynes M. Proteomic analysis of multidrug-resistance mechanisms in adriamycin-resistant variants of DLKP, a squamous lung cancer cell line. Proteomics. 2009;9:1556–1566. doi: 10.1002/pmic.200800633. [DOI] [PubMed] [Google Scholar]


