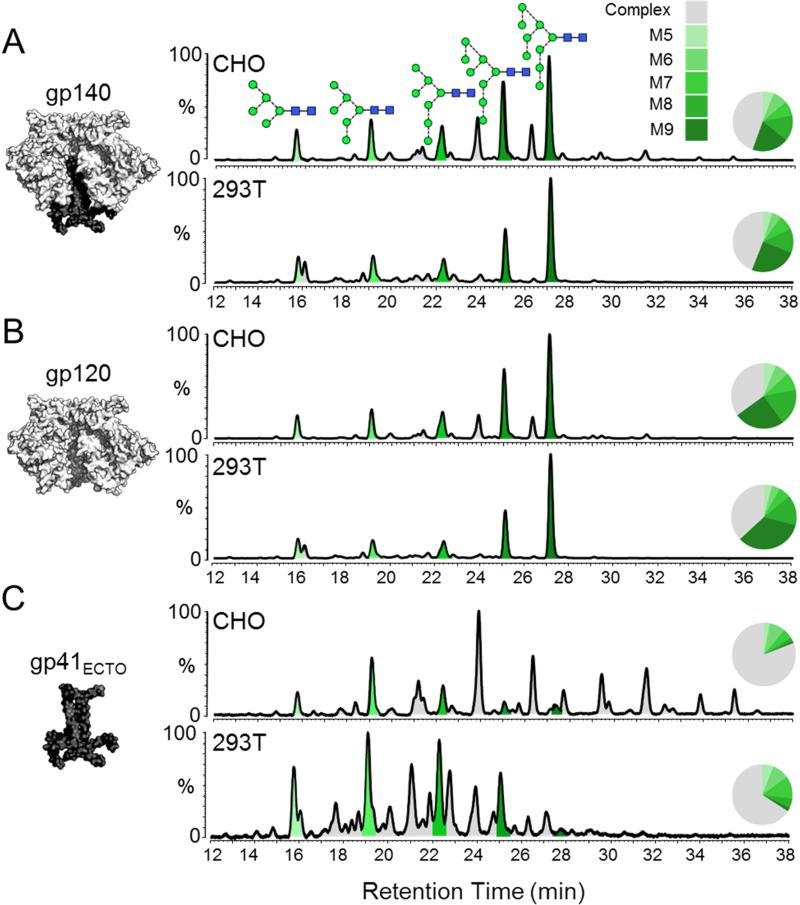
Figure 1. Comparison of glycosylation patterns of BG505 SOSIP.664 trimers produced in CHO and 293T cells.
HILIC-UPLC spectra of fluorescently labelled N-linked glycans isolated from (A) the entire trimeric gp120/gp41ECTO complex (gp140), and the (B) gp120 and (C) gp41ECTO subunits of BG505 SOSIP.664 trimers. The subunits analyzed in each case are illustrated on the left. Trimers were purified by 2G12-affinity chromatography followed by SEC. The gp140 band was extracted from a non-reducing SDS-PAGE gel, while the gp120 and gp41ECTO bands were resolved by SDS-PAGE under reducing conditions. The glycan contents of proteins extracted from the bands were analyzed. Peaks corresponding to oligomannose glycans (M5-M9; Man5-9GlcNAc2) are colored in shades of green; the remaining peaks (gray) correspond to complex and hybrid-type glycans. Peak areas (as a % of total glycans) are illustrated in the associated pie charts. Glycan structures are represented according to the color scheme established by the Consortium for Functional Glycomics (http://www.functionalglycomics.org/). The SOSIP.664 construct contains the following mutations: a T332N mutation to introduce the 332 glycosylation site; cysteines at 501 and 605 form a disulphide bridge to covalently link the gp120 and gp41ECTO subunits; replacement of the gp120 furin cleavage site (RXXR) with a hexa-arginine (R6) sequence to promote furin cleavage; an I559P mutation to stabilize the gp41ECTO subunits in the pre-fusion form; and deletion of the MPER region at residue 664 to reduce aggregation.