Fig. 5.
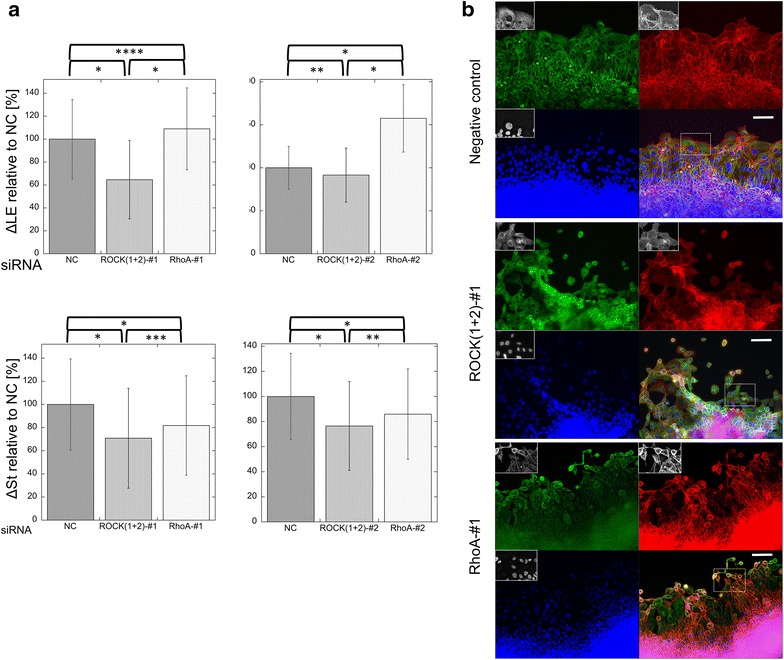
Migration of TE-10 cell sheets with siRNA knockdown of target mRNAs 72 h after scraping. a Cell migration was quantified over a 48-h period. For each experiment, negative control values were used as the reference. The upper graphs show the migration velocity of the leading edge (Δ LE), while the lower graphs show the velocity of the leading line of the stratified region. Data shown represent the mean ± SEM of at least six independent experiments in which at least three series were stained with Hoechst 33342 and at least 3 series were stained with H2B-GFP. *p < 0.001; **p = 0.002; ***p = 0.0028; ****p = 0.0044. ANOVA was performed using the original data. NC negative control. b Images of the cell sheets 72 h after scraping under siRNA treatment. First row negative control scrambled siRNA. The shape of the cells in the front row was very similar to that of untreated cells. There was no obvious irregularity in the arrangement of the cells. Second row knockdown of ROCKs by siRNA (ROCK1-#1 and ROCK2-#1). The arrangement of the cells in the leading row was obviously disordered. Many cells in the leading row were small with hypoplastic stress fibers. Several empty spaces between the cells were visible. Third row knockdown of RhoA by siRNA (RhoA-#1). The cells in the simple layer region were smaller than those in the negative control. Many of the cells were small and round, with inconspicuous stress fibers. Green α-tubulin, red β-actin, blue nuclei; scale bar 100 µm. Each inset shows the magnified image of the area surrounded by the interrupted white line on the merged image
