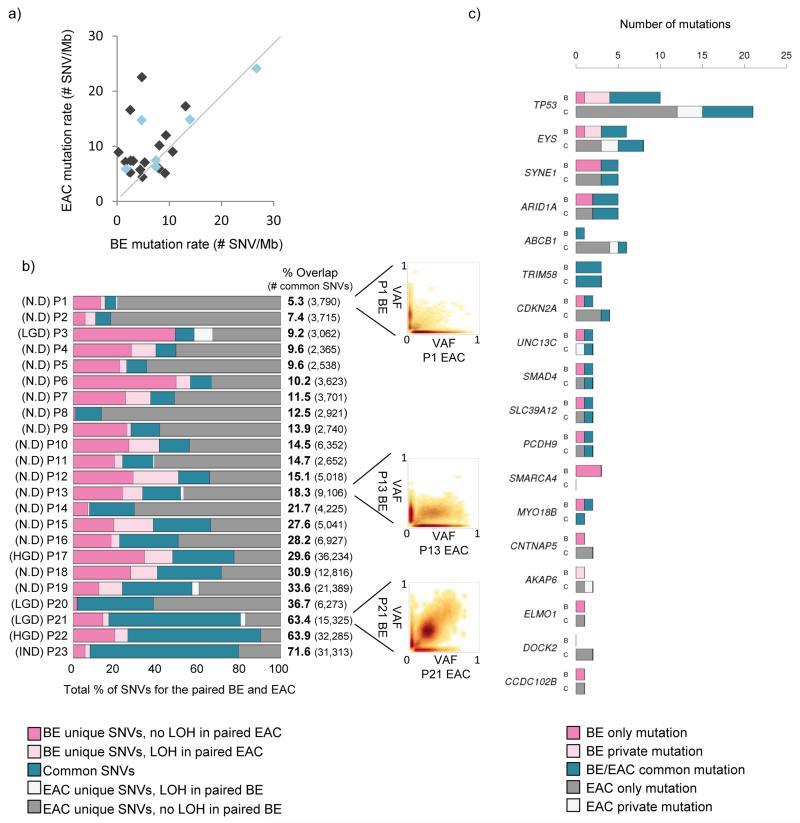
Figure 1. Paired Barrett’s and EAC samples have a varied overlap.
a) Scatter plot comparing the mutation rate between paired Barrett’s and EAC samples. Light blue dots indicate Barrett’s samples with some degree of dysplasia present. b) Diagram showing percentage overlap between SNVs in paired Barrett’s and EAC samples, including highlighting SNVs that lie in areas of LOH in the reciprocal sample. Barrett’s unique SNVs that lie in an area of LOH in the paired EAC sample are shown in light pink and EAC unique SNVs that lie in a region of LOH in the paired Barrett’s sample are shown in white. Samples are ranked according to their degree of overlap, from poor to good. Scatter plots illustrating an example of a poor overlap (patient P1), a fair overlap (patient P13) and a good overlap (patient P21) are shown. c) Bar graph showing genes that were found to be recurrently mutated in previous EAC sequencing studies13,16 and are mutated in at least two patients in our Barrett’s-EAC cohort (SNVs or indels). For each gene of interest the data for the Barrett’s samples are in the bar labelled “B” and for cancer samples in the bar labelled “C”. Mutations that are common to both paired Barrett’s and EAC samples (shown in teal), as well as mutations that are Barrett’s unique (dark pink) or EAC unique (dark grey) are shown. Also shown are mutations in the same gene that have a different base pair change between paired Barrett’s and EAC samples (called “private mutations”).