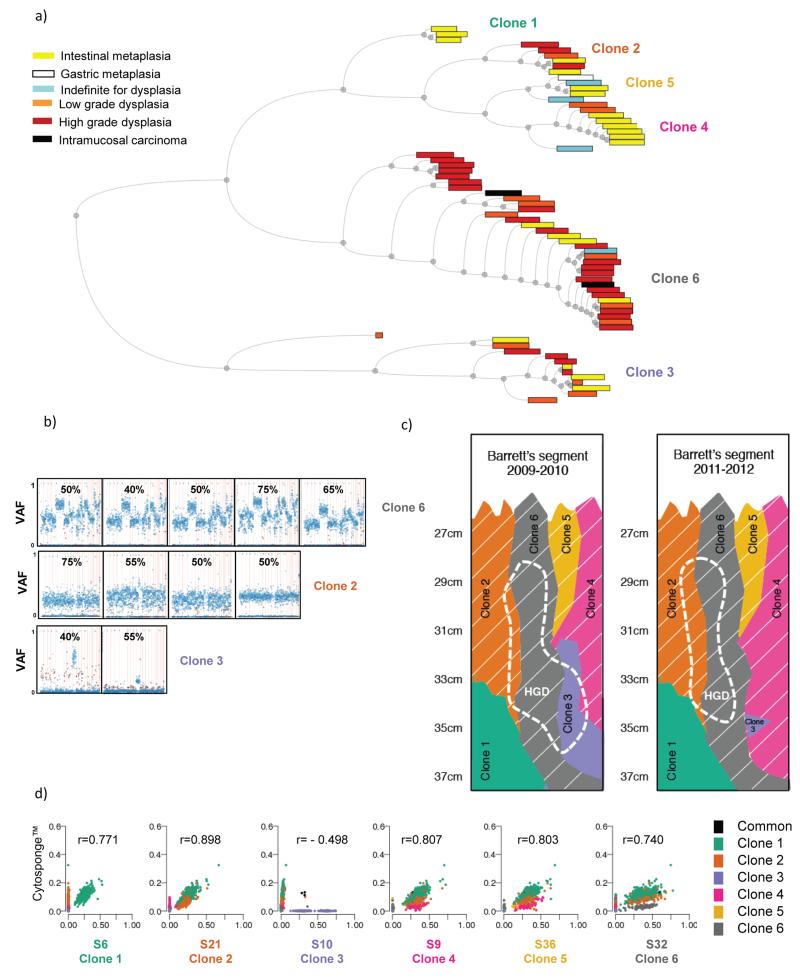
Figure 6. Multiple different clones can give rise to dysplasia.
a) Hierarchical tree showing all samples processed for amplicon re-sequencing. Within the tree, samples are paired according to their Pearson correlation score. The distance between pairs is represented by the horizontal distance between the pair and their parent node. There is no information embedded in the vertical distance of this tree. The colors of the boxes represent the histopathological grade for each of the Barrett’s esophagus samples. The width of the boxes is proportional to the number of SNVs with VAF>0.01 in each sample. The clones represented by the six different branches are noted. b) VAF plots as described in Figure 5a showing examples of samples that contain ≥40% high grade dysplasia (as determined by the average between two expert upper gastrointestinal pathologists) within the sample and grouped according to which clones they represent. The number at the top of each graph indicates the percentage of high grade dysplasia in that specific sample, as assessed by two expert gastrointestinal pathologists. c) Illustration depicting the clonal arrangement in patient AHM1051’s Barrett’s esophagus segment before (2009-2010) and after (2011-2012) endoscopic treatment. The region corresponding to high grade dysplasia, before and after treatment, is shown using a dashed line. The clones containing the widely-spread TP53 mutation are indicated by thin, slanted, white lines. d) Scatter plots indicating the correlation between SNVs identified in the DNA from cells collected using the Cytosponge compared with the six different clones. The r value represents the Pearson correlation.