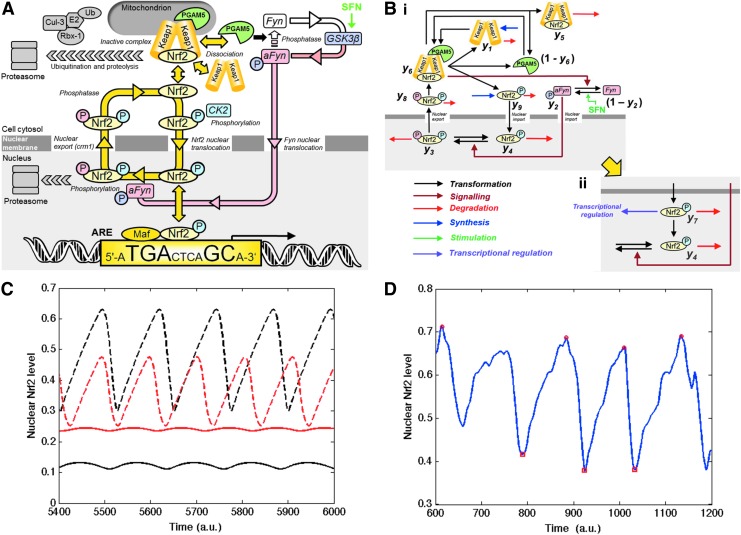
FIG. 5.
Cell signaling sustaining Nrf2 translocational oscillations for control of ARE-linked gene expression. (A) Schematic diagram. (B) Regulatory model—(i) model of the signaling network and (ii) extended model with inactivation of nuclear Nrf2 introduced. Negative feedback is provided by successive de-phosphorylation and phosphorylation steps, which induce ultra-sensitivity in Fyn activation and the Fyn phosphorylation of nuclear Nrf2 and which produce a delay between Nrf2 nuclear entry and the Fyn phosphorylation which drives nuclear export. Variables y1−y9 are indicated. (C) Simulations of Nrf2 oscillation in the extended model. Two levels of Nrf2 in the nucleus, total (Nrf2+iNrf2+pNrf2, normalized) and (Nrf2, non-normalized), are calculated in the control and SFN-stimulated conditions, which are modeled respectively by small and large values of the parameter (h1) that represents the maximum rate of Fyn activation. Key: (—) active nuclear Nrf2, (- - -) total nuclear Nrf2, h1=0.168; (solid red lines) active nuclear Nrf2, and (dashed red lines) total nuclear Nrf2, h1=0.264. See Supplementary Data for other parameter values and equations. Active nuclear Nrf2 is non-normalized and total nuclear Nrf2 is normalized. (D) Stochastic model in the control condition. The result is smoothed by wavelets in the same way as used in the processing of the video frames. The time scale is given in arbitrary units (a.u.). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars