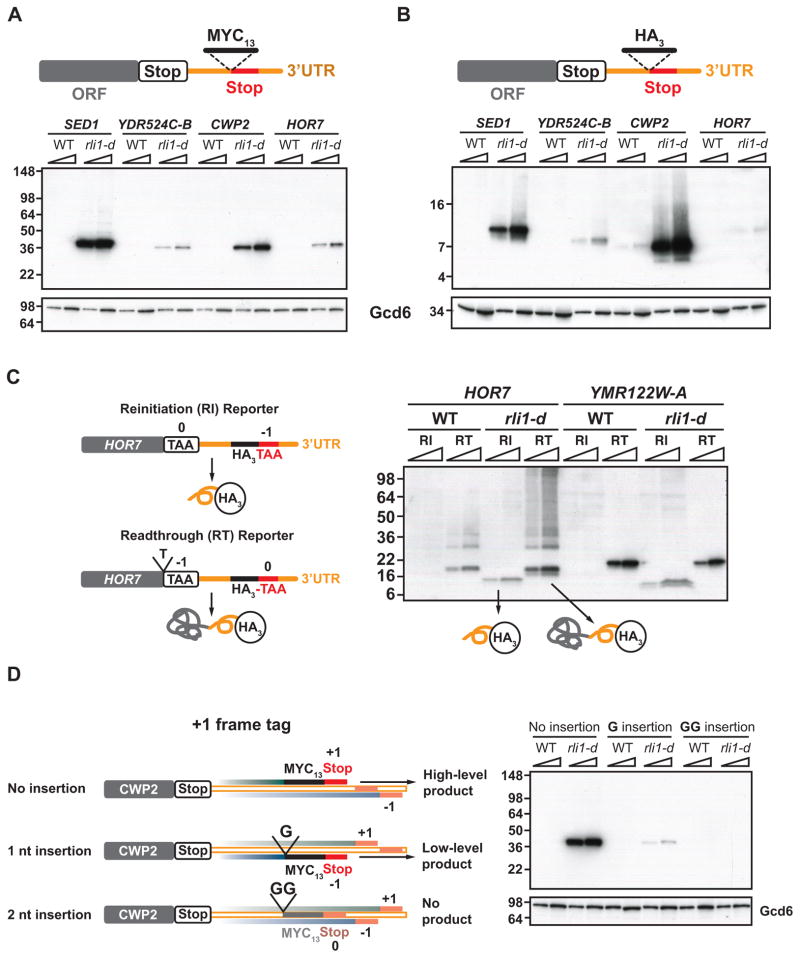
Figure 4. Detection of epitope-tagged 3′UTR translation products that result from reinitiation versus proteolysis of readthrough products in rli1-d cells.
(A–B) MYC13 (A) or HA3 (B) epitope tags were inserted just upstream of predicted 3′UTR termination sites in the chromosomal alleles of 4 candidate genes (depicted schematically) in WT and rli1-d strains. Tagged strains were cultured as in Fig. S1A and WCEs were subjected to Western analysis using antibodies against c-Myc or HA (upper blots) or Gcd6 (lower blots). Two amounts of extracts were loaded in each lane pair. Migration of MW standards is indicated on the left. ORF, main coding sequences. (C) HA3-tagged reinitiation (RI) and readthrough (RT) reporters for HOR7, and their predicted tagged products, are depicted schematically on the left. The RI reporters for HOR7 and YMR122W-A were those analyzed in (B), and the RT reporters were constructed from them by inserting a single nucleotide (T for HOR7) before the main ORF stop codon, putting that terminator out of frame, and placing the 3′UTR termination site in frame with the main ORF. Western analysis of the resulting strains was conducted as in (B). (D) To detect CWP2 3′UTR translation in different reading frames, single or tandem G nucleotides were inserted immediately preceding the MYC13 tag located adjacent to the predicted 3′UTR +1-frame termination site in the CWP2 reinitiation reporter analyzed in (A) and depicted here as “No insertion”, thus shifting the MYC13 tag and adjoining in-frame stop codon from the +1 frame into the −1 or 0 frames for the 1 nt and 2 nt insertions, respectively, as depicted schematically. The first stop codons encountered downstream of the tag coding sequences in the +1 (top), 0 (middle), and −1 (bottom) frames are indicated with filled red or orange boxes. For the 2 nt insertion/GG construct, the tag is shown partially transparent and the adjoining stop codon in orange versus red to indicate the absence of detectable reinitiation in the 0 frame. Tagged WT and rli1-d cells were analyzed as in (A).