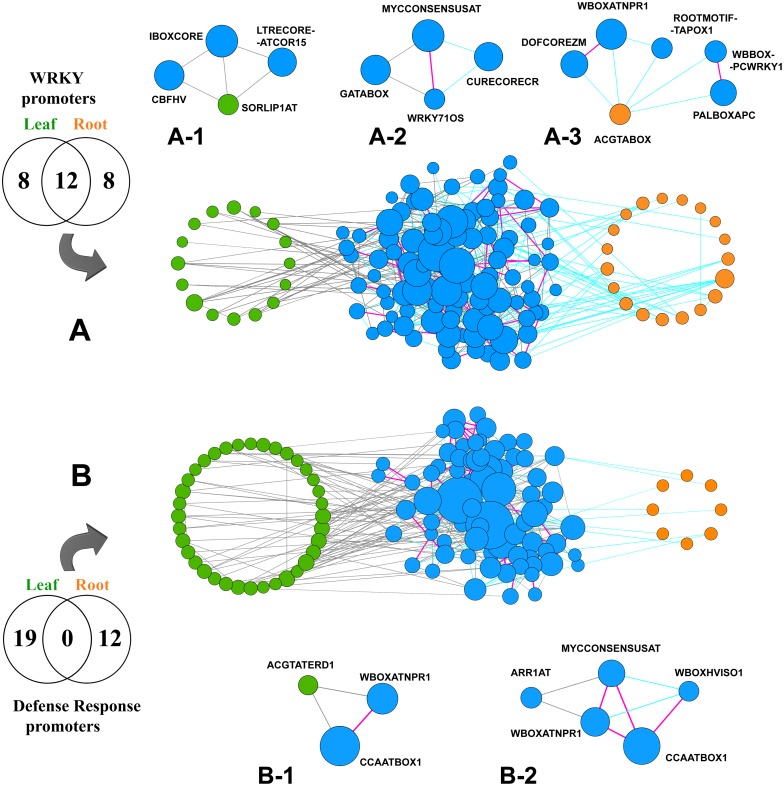
Fig 6. Co-occurrence networks of two pairs of input gene-sets (derived from 4 DPI leaf and 2 DPI root data) and their respective cliques are presented.
(A) The CRE co-occurrence networks computed for the up-regulated WRKY genes in rice leaf and root tissues are merged into a single one. Nodes (individual CREs) explicitly found in the leaf are highlighted (green) in the left, while those found in the root are highlighted (orange) in the right hand side of the network. Nodes in the middle (blue) are found in both leaf and root tissues. Gray and cyan colour of edges imply that a significant co-occurrence is found between the two connected CREs explicitly in leaf and root respectively. A pink edge implies that the two connected CREs significantly co-occur in both the tissues. (A-1) An example of two cliques [SORLIP1AT, CBFHV, IBOXCORE] and [SORLIP1AT, IBOXCORE, LTRECOREATCOR15], explicitly occurred in the network of leaf. (A-2) An example of two cliques, found in leaf and root respectively, where they share a common edge (WRKY71OS—MYCCONSENSUSAT). (A-3) An example of three cliques [ACGTABOX, DOFCOREZM, WBOXATNPR1], [ACGTABOX, WBOXATNPR1, ROOTMOTIFTAPOX1] and [ACGTABOX, PALBOXAPC, WBBOXPCWRKY1], out of which one clique explicitly occurred in root. (B) The merged CRE co-occurrence networks of rice leaf and root, computed for the “Defense Response” gene-sets. (B-1) An example of a clique, occurred in the network of leaf. (B-2) An example of four cliques, out of which one occurred in the network of leaf, two occurred in the network of root and one is common [CCAATBOX1, WBOXATNPR1, MYCCONSENSUSAT] in both the networks.