Figure 1.
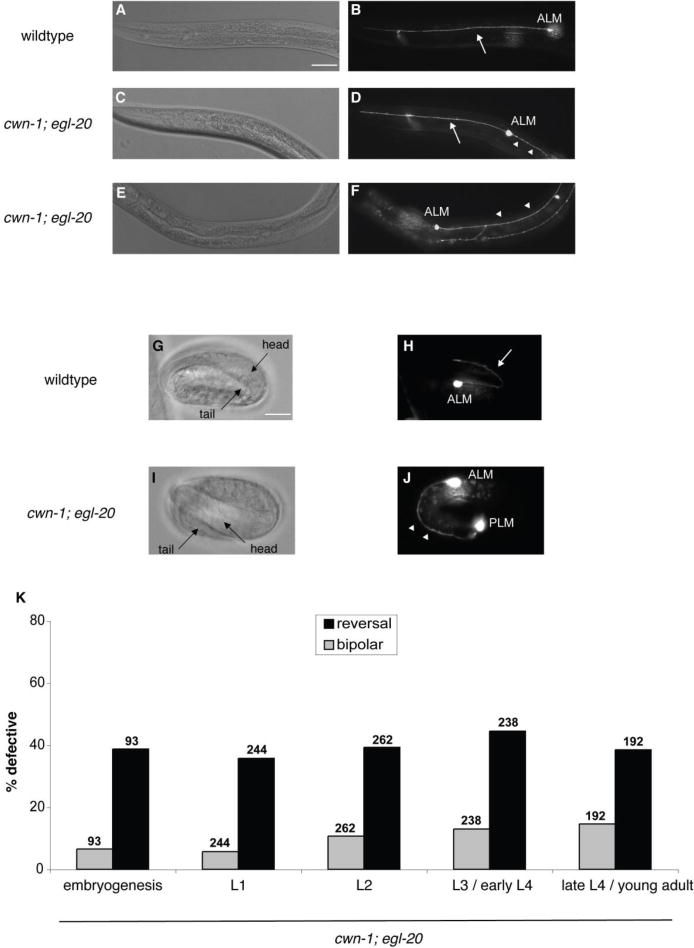
Wnts orient the initial polarity of ALM. (A, C, E) Nomarski and (B, D, F) fluorescence photomicrographs showing the anterior of fourth larval stage zdIs5 [Pmec-4::gfp] animals. (A, B) A wild-type larva. (C, D, E, F) cwn-1; egl-20 mutant larvae. The ALM cell bodies are labeled. Arrows indicate anterior processes, and arrowheads indicate posterior processes. Anterior is to the left, dorsal is up. Scale bar represents 30 μm. (B) A wild-type ALM extends a single, anterior process. (D) A cwn-1; egl-20 ALM is bipolar, extending both a normal anterior process and an ectopic posterior process. (F) A cwn-1; egl-20 ALM extends a single posterior process, indicating a reversal of polarity. (G–J) Photomicrographs of zdIs5 embryos showing ALM neuronal morphology. G and I are Nomarski images of H and J, respectively. (G, H) A wild-type embryo. (I, J) A cwn-1; egl-20 mutant embryo. Scale bar represents 10 μm. (H) A wild-type 3-fold embryo extends a normal anterior process. (J) A cwn-1; egl-20 3-fold embryo extends a single posterior process toward the tail, indicating a reversal of polarity. (K) Graph shows the percentage of ALM neurons with defective polarity for cwn-1; egl-20 animals at different developmental stages. Gray bars indicate the bipolar phenotype and black bars indicate reversed polarity. The number of ALMs scored is indicated above the bars for each genotype. L1, L2, L3 and L4 are the first, second, third and fourth larval stages, respectively. cwn-1; egl-20 animals at different stages displayed comparable ALM reversal defects. We found significant differences between the number of bipolar ALMs in L1 and L3 / early L4 animals and between L1 and late L4 / young adult animals (p < 0.001; Fisher’s exact test). We believe this difference is due to a technicality in how the numbers of bipolar neurons are scored (see Results).
