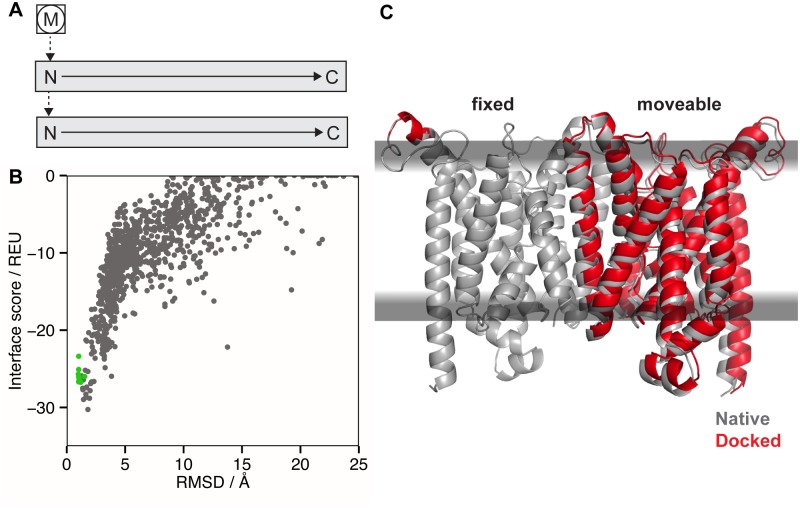
Fig 7. Protein-protein docking in the membrane bilayer using MPdock.
(A) FoldTree representation used in MPdock with the membrane residue (M) being fixed at the root (circled) of the FoldTree and the protein chains as docking partners attached via jump edges. (B) Interface score vs. backbone RMSD to the native structure for 1000 models of the vitamin B12 importer BtuCD. The RMSD is the ‘ligand’ RMSD, which is computed only over the moving partner after superimposing the fixed partner and membrane. The green dots represent ten models created by minimizing the crystal structure. The interface score of the crystal structure (180.5 REU) is outside of the plotting range due to clashes. (C) Native structure of the vitamin B12 importer (gray, PDB 2qi9) superimposed with the model having the lowest interface score (red).