FIG 3 .
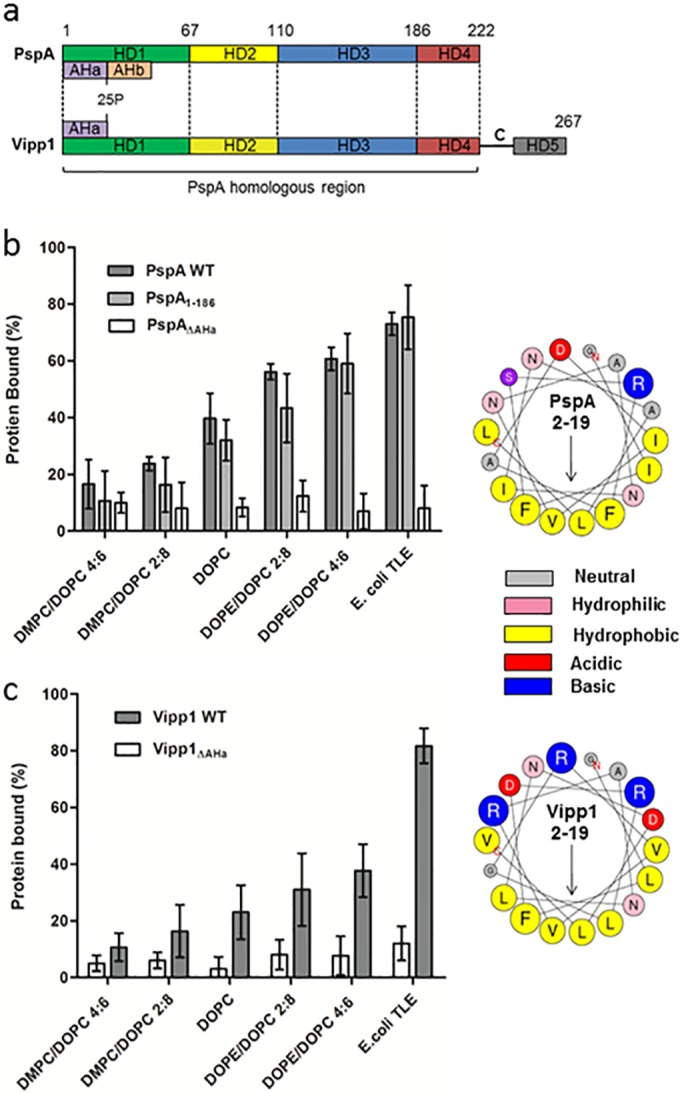
Membrane binding of PspA variants and the PspA homologue Vipp1. (a) Schematic of the E. coli PspA and Synechocystis Vipp1 protein sequences, with the positions of the helical domains (HD1 to 4 according to regions defined for PspA and HD5 of Vipp1) and AH regions. (b) Affinity of PspA, PspA1–186, and PspAΔAHa for vesicles under increasing SCE stress and E. coli TLE (1 mM lipid). (c) Affinities of Vipp1 and Vipp1ΔAHa within the same assay but at a 2 mM lipid concentration. Concentrations for all proteins were 10 µM within the assay. ANOVA showed differences in binding between lipid composition to be statistical significant (P < 0.05) for all proteins apart from the ΔAHa mutants of PspA and Vipp1. (b and c) Right sides show helical wheel projections of the N-terminal AHa (residues 2 to 19) of PspA and Vipp1, with arrows showing the direction of the hydrophobic moment. Residue sizes are proportional to the amino acid side chain volumes.
