Abstract
Tumor-forming endothelial cells have highly elevated levels of Nox-4 that release H2O2 into the nucleus, which is generally not compatible with cell survival. We sought to identify compensatory mechanisms that enable tumor-forming endothelial cells to survive and proliferate under these conditions. Ape-1/ref-1 (Apex-1) is a multifunctional protein that promotes DNA binding of redox-sensitive transcription factors, such as AP-1, and repairs oxidative DNA damage. A validated mouse endothelial cell (EOMA) tumor model was used to demonstrate that Nox-4-derived H2O2 causes DNA oxidation that induces Apex-1 expression. Apex-1 functions as a chaperone to keep transcription factors in a reduced state. In EOMA cells Apex-1 enables AP-1 binding to the monocyte chemoattractant protein-1 (mcp-1) promoter and expression of that protein is required for endothelial cell tumor formation. Intraperitoneal injection of the small molecule inhibitor E3330, which specifically targets Apex-1 redox-sensitive functions, resulted in a 50% decrease in tumor volume compared with mice injected with vehicle control (n = 6 per group), indicating that endothelial cell tumor proliferation is dependent on Apex-1 expression. These are the first reported results to establish Nox-4 induction of Apex-1 as a mechanism promoting endothelial cell tumor formation.
Keywords: hemangioendothelioma; hemangioma; Nox-4; AP-1 transcription factor; c-Jun transcription factor; endothelial cell; tumor cell biology; DNA damage response; monocyte chemoattractant protein-1, Ape-1/ref-1
endothelial cell (EC) tumors are the most common soft tissue tumor of infancy frequently resulting in significant deformity and sometimes even death (20, 22, 36). Although many of these tumors resolve spontaneously, 10% are considered “threatening,” e.g., potential ear, eye, nasal, oral, or airway obstruction, and 1% threaten the life of the child. The majority of these tumors occur in the head and neck, causing the patients and families to live with a visible deformity. Treatment is limited to those with threatening or life-threatening tumors because of the high-risk side-effect profile associated with the current pharmacological agents being used. However, over 50% of children have residual deformity when the tumors involute, including those that receive treatment (56). A better understanding of the mechanisms that promote EC tumor growth will facilitate the development of new treatment interventions for children with EC tumors, so that all children, not just those with threatening tumors, can be treated and their deformities prevented.
Previous work using EOMA cells as a validated mouse model of EC tumors reported significantly elevated levels of Nox-4, the catalytic subunit of NADPH oxidase (21, 58). H2O2, the biologically active oxidant species derived from Nox-4, was present in significantly higher levels in EOMA cells compared with non-tumor-forming murine aortic endothelial (MAE) cells and was expressed directly into the nucleus of EOMA cells, resulting in elevated levels of oxidized DNA (8-OHdG) present in the urine of tumor-bearing mice (14). Oxidant-inducible monocyte chemoattractant protein-1 (MCP-1) expression is required for EC tumor formation, and Nox-4 and oxidant-sensitive AP-1 activation are essential upstream mediators of MCP-1 protein expression in EOMA cells (13, 17). The EOMA model was used to show that EC tumor growth is Nox-4 dependent and that antioxidant interventions can significantly inhibit EC tumor growth (1, 14). These works from our laboratory have established that the Nox-4/MCP-1 signaling pathway is required for EC tumor formation.
The Nox-4/MCP-1 signaling pathway is implicated in EC tumorigenesis because NADPH is the predominant source of reactive oxygen species (ROS) production in ECs and the widespread recognition that oxidative stress observed in many cancers likely plays an etiologic role (6, 28–29). AP-1 is an oxidant-inducible transcription factor with multiple mcp-1 promoter binding sites (19), and when AP-1 is constitutively activated it has been shown to promote tumor formation (41, 44, 55). NADPH is a constitutively active protein with pathologically elevated levels in EOMA cells (14). Increased expression of MCP-1 is required for angiogenesis that is inherent to the growth of many different solid tumors, including those of EC origin (17, 18, 51,52). One of the objectives of this work was to establish a link between elevated Nox-4 expression levels, AP-1 activation, and EC tumor growth. However, the previously reported finding that Nox-4-derived H2O2 was expressed into the nucleus challenged that hypothesis because a dominant oxidizing environment in the nucleus is known to generally inhibit transcription factor activation (7, 48).
To test the hypothesis that Nox-4-induced expression of AP-1 promoted EC tumor formation, the compensatory mechanisms that enable AP-1 to bind to its cognate site in the mcp-1 promoter despite excessive Nox-4-derived nuclear H2O2 were also investigated. Ape-1/ref-1 (Apex-1) is a multifunctional protein that regulates DNA binding of several redox-sensitive transcription factors, including AP-1, by maintaining the cysteine residues at their DNA binding sites in a reduced state (4). In this work we test the hypothesis that Nox-4-derived H2O2 not only induces AP-1 transcriptional activity, but also coinduces Apex-1 protein expression to enhance AP-1-dependent MCP-1 expression. Furthermore, induction of Apex-1 mitigates the deleterious effects of DNA damage induced by pathologically elevated levels of Nox-4. We posit that pathologically induced expression of Apex-1 represents a compensatory mechanism that promotes survival of tumor-forming ECs and enables the Nox-4/MCP-1 axis to drive EC tumor growth in vivo.
MATERIALS AND METHODS
Materials
The following materials were obtained from the source indicated: hexadimethrine bromide, dimethyl sulfoxide, E3330 (Sigma, St. Louis, MO) and calcein AM (Invitrogen, Grand Island, NY). For cell culture, Dulbecco's modified Eagle's medium (DMEM), fetal calf serum (FCS), and penicillin and streptomycin were purchased from Invitrogen (Invitrogen). Culture dishes were obtained from Nunc (Nunc, Rochester, NY). MCP-1 promoter vector was a gift of Dr. Brad Rovin, Ohio State Univ., and PF6-AM H2O2 fluorescent detection probe was a gift of Prof. Christopher J. Chang and Bryan C. Dickinson, Univ. of California, Berkeley.
Methods
Cell culture.
Murine endothelial (EOMA) cells were maintained under the same conditions as previously described (15). In brief, EOMA cells were maintained in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin (complete media) and incubated at 37°C and 5% CO2.
In vivo studies.
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Ohio State University, Columbus, OH. Mice were maintained under standard conditions at 22 + 2°C with 12:12-h dark-light cycles with access to food and water ad libitum. 129P/3 mice (6–8 wk, female, Jackson Laboratories, Bar Harbor, ME) were subcutaneously injected with EOMA cells, as previously described (15). Tumor volume was determined by using calipers to measure length × width × height of each tumor, and the mass was determined by draining the blood from the tumor and weighing the residual solid tumor mass dissected free from any surrounding soft tissue as previously described (14–15, 17). At the time of EOMA cell injection, E3330 (25 mg/kg) and CRT0044876 (5 mg/kg) was injected intraperitoneally, twice daily with 8 h interval for 7 days either alone or in combination. Mice were euthanized 7 days after EOMA cell injection, and tissues were collected frozen in OCT compound for histological analyses and liquid nitrogen for RNA isolation.
Determination of cell viability.
Cell viability was measured by extracellular leakage of lactate dehydrogenase per manufacturer's instructions (Sigma Chemical, St. Louis, MO). Cell viability was also determined by incubating cells with calcein-AM (3 mmol/l) and propidium iodide (PI) (2.5 mmol/l) in phosphate-buffered saline for 15 min at 37°C and with 5% CO2. Digital images were collected using Zeiss Axiovert 200M microscope as described previously (25, 26, 39) Fluorescence intensity was also determined by FACs using PI staining in the FL2 region using an Accuri C6 Flow Cytometer (Accuri, Ann Arbor, MI) at 530-nm excitation with a gated sample size of 10,000 cells. (43).
Cell proliferation assay.
EOMA cells were seeded in 12-well plates at 1 × 105 cells/well for 14–18 h. Cell proliferation was analyzed using Cell Proliferation ELISA, BrdU kit (Roche, Basel, Switzerland). BrdU was added to a final concentration of 1 μM. After incubation for further 24 h, DNA synthesis was assayed using colorimetric technique as per manufacturer's protocol.
Western blot.
Immunoblotting was performed using EOMA cell lysates and the protein concentration determined using a BCA protein assay. Samples (25–40 μg of protein/lane) were separated on a 4–12% SDS polyacrylamide gel electrophoresis and probed with rabbit monoclonal anti-Nox-4 antibody (1:1,000 dilution, Epitomics, Burlingame, CA), rabbit polyclonal anti-Apex-1 antibody (1:500 dilution, Abcam, Cambridge, MA), and anti-mouse β-actin (1:10,000 dilution, Sigma). Bands were visualized by using horseradish peroxidase-conjugated donkey anti-rabbit-IgG (Amersham Biosciences, Piscataway, NJ, cat. no. NA934V) at 1:2,000 dilution and the enhanced chemiluminescence assay (Amersham Biosciences), according to the manufacturer's instructions. Pixel densitometry for individual bands was done using ImageJ software.
MCP-1 and 8-OHdG ELISA.
EOMA cells were seeded in 12-well plates at 1 × 105 cells/well. MCP-1 ELISA was performed on cell supernatants by using the CCL2/JE/MCP-1 Quantikine ELISA kit (R&D Systems, Minneapolis, MN) and 8-OHdG ELISA kit (JaICA, Japan) (27) according to the manufacturer's instructions. BCA protein assay was performed on an aliquot of all tested samples and the results standardized per milligram of protein.
c-Jun DNA binding assay.
c-Jun DNA binding activity was assessed using the Trans-Am AP-1 Transcription Factor ELISA Kit (Active Motif, Carlsbad, CA). EOMA cells were plated in 100-mm tissue culture dishes at 2 × 106 cells/plate. After specified time indicated in figure, cells were harvested by scraping and nuclear extracts isolated using the Active Motif nuclear isolation kit (Active Motif). Nuclear extract was quantified using the DC protein reagent kit (Bio-Rad Hercules, CA). A 2-μg aliquot of nuclear extract was placed in the wells of a 96-well plate followed by incubation with primary and secondary antibodies sequentially. The primary antibody detects activated/phosphorylated c-Jun by recognizing an epitope on c-Jun that is accessible only upon DNA binding. A horseradish peroxidase-conjugated secondary antibody was used for colorimetric detection of the bound primary antibody. Wild-type consensus oligonucleotide that competes for the AP-1 binding site was used as a control to confirm the specificity of the assay (data not shown). Absorbance at 450 nm was used for detection.
Immunohistochemistry and immunocytochemistry.
Immunohistochemistry of Nox-4 was performed on cryosections from human and mouse tumor samples using specific antibody as described previously (17). In brief, murine tissue specimens were snap frozen in optimum cutting temperature (OCT) compound (Miles, Elkhart, IN). Frozen tissues were sectioned at 10–12 μm thickness, fixed for 5 min in acetone at 4°C, and stained using DAB peroxidase (HRP) substrate kit, 3,3′-diaminobenzidine (Vector Laboratories, Burlingame, CA). Anti-rabbit Nox-4 monoclonal antibody (Epitomics, Burlingame, CA) was used at 1:100 dilution. Negative control sections were stained using biotinylated goat anti-rabbit secondary antibody to test primary antibody specificity. Tissue sections were counterstained with hematoxylin. Images were obtained under 20× magnification, and intensity (Nox-4, brown) quantitation was performed using the auto measure tool in Axiovision 4.6 software, (Axiovert 200M; Zeiss, Germany). All human tissue sections were formalin-fixed, paraffin-embedded archival tissue specimens. Immunohistochemical staining of paraffin or frozen sections was performed using the following primary antibodies: Nox-4 (1:100), c-Jun (1:100), phospho c-Jun (1:50, Cell Signaling, Danvers, MA), and 8-OHdG (1:50). After heat-induced epitope retrieval for 20 min, the sections were permeabilized using a buffer (0.2% Triton X-100 in PBS) for 10 min. For detecting phospho-c-Jun sections are treated with 1 μM okadaic acid (Sigma) for 1 h after the permeabilization step. Secondary antibody detection and counterstaining were performed as described (10). For immunocytochemistry, cells (1 × 105 cells/well) were seeded on a coverslip placed in 12-well plates for 24 h, then transfected with siRNA oligonucleotides. Seventy-two hours posttransfection, cells were washed with PBS thrice and then fixed, permeabilized, and blocked for nonspecific signal. Next, the cells were incubated with primary antibody with dilutions indicated in parentheses: Nox-4 (1:100), Apex-1 (1:100), and 8-OHdG (1:50). The cells were washed and incubated with Alexa-flour 488 goat anti-rabbit antibody (1:200 dilution, Invitrogen) and Alexa-flour 568 goat anti-rabbit antibody (1:200 dilution, Invitrogen) for 1 h at room temperature. After incubation, the cells were mounted in gelmount containing DAPI (aqueous mount, Vector Laboratories). The images were captured by confocal microscope and the quantification of fluorescent intensity was measured using FV10-ASW 3.0 software (Olympus, Tokyo).
Cell fractionation.
EOMA and MAE cells (1 × 105 cells/12 well plate) were cultured for 24 h. Cells were washed twice with PBS and harvested using cell lifter. Subcellular fractionation of cells was performed using the Qproteome cell compartment kit (Qiagen, Valencia, CA) by following the manufacturer's directions. Purity of the nuclear, cytoplasmic, and mitochondrial fractions was confirmed by measuring nuclear, cytoplasmic, and mitochondrial membrane-specific protein markers such as p84 (1:5,000; GeneTex, Irvine, CA), β-actin (1:10,000; Sigma), and cytochrome c (1:2,000; BD Pharmingen, San Diego, CA) with Western blot assay by following the manufacturer's procedures.
Transfection experiments and analysis of gene expression.
The cells were seeded in a 12-well plate at a density of 0.1 × 106 cells/well for 24 h before treatment. Delivery of siRNAs was achieved using DharmaFECT 1 transfection reagent (Thermo Fisher Scientific, Lafayette, CO). All siRNA smart pool was obtained from Dharmacon RNA Technologies. For controls, siControl non-targeting siRNA pool (mixture of 4 siRNA, designed to have ≥4 mismatches with the corresponding gene) was used (53). DharmaFECT 1 Transfection Reagent (Dharmacon RNA Technologies, Lafayette, CO) was used to transfect cells with mentioned siRNAs (Dharmacon RNATechnologies, Lafayette, CO) for 72 h as per the manufacturer's instructions. Unless specified, the cells were lysed after 72 h of transfection, and RNA/protein were collected for gene expression study (24–26, 39). For detection of mRNA, total RNA from cells was extracted using miRVana miRNA isolation kit according to the manufacturer's protocol (Ambion, Life Technologies, Grand Island, NY). For mRNA expression studies, cDNA synthesis was achieved by a SuperScript III first-strand synthesis system (Applied Biosystems, Life Technologies) as described (16). The abundance of mRNA for genes of interest was quantified by using real-time polymerase chain reaction (PCR) with double-stranded DNA binding dye SYBR green-I. The following forward (F) and reverse (R) primer sets (Invitrogen) were used: M_JE/MCP-1 F: 5′-GAA GCT GTA GTT TTT GTC ACC AAG C-3′; M_JE/MCP-1 R: 5′-GAT CTC ATT TGG TTC CGA TCC AG-3′; M_18s rRNA F: 5′-GTA ACC CGT TGA ACC CCA TT-3′; M_18s rRNA R: 5′- CCA TCC AAT CGG TAG CG-3′; M_Nox-4 F: 5′-CAC CTC TGC CTG CTC ATT TGG-3′; M_Nox-4 R: 5′-AGT TGA GGT TCA GGA CAG ATG CA-3′; M_Apex 1 F: 5′-CTT CGT TGG GAG GCA GCG CA-3′; M_Apex 1 R: 5′-CTC TCC CGCGGC CTC CTT CT-3′.
Cellular H2O2 analysis.
To determine intracellular H2O2 levels, EOMA cells were incubated with 5 mM PF6-AM in PBS for 20 min at 37°C temperature as described (3, 32, 38). To limit oxidative detection of peroxynitrite, cells were pretreated with l-NAME (10 μM) for 1 h prior to addition of PF6-AM. To detect the specific level of H2O2 generation in different compartments of EOMA cells, different organelle-specific dyes were used. For nuclear staining Hoechst (H) 33342 (1 μM, Molecular Probes, Grand Island, NY) and for mitochondrial staining, MitoTracker (MT) Red (0.25 μM, Molecular Probes) were used in the media for 20 min at 37°C along with PF6-AM. After loading, cells were washed twice to remove excess dye. The cells were imaged using a Zeiss Axiovert 200M microscope. The quantification of fluorescent intensity was measured using auto measure mode of Axivision software (Zeiss, Germany).
Target reporter luciferase assay.
EOMA cells (0.075 × 106 cells/well in 12-well plate) were seeded in antibiotic-free medium for 18–24 h prior to transfection. Cells were transfected with control oligonucleotides and nox4/apex-1 siRNA for 72 h (39). After 72 h, cells were retransfected with 500 ng of pAP1-Luc (Agilent technologies, Santa Clara, CA) or mcp-1 promoter plasmid using Lipofectamine LTX PLUS reagent (Invitrogen). Luciferase activity was determined using the luciferase reporter assay system (Promega, Madison, WI).
Oxidative DNA isolation and transfection.
Both EOMA/MAE cells were trypsinized (6–7 × 106 cells), followed by resuspending in 3 ml of TE (Tris-EDTA) containing 20% vol/vol SDS (100 μl). After 1 h, proteinase K (20 mg/mL, 20 μl) was added by mixing gently by inversion followed by incubation overnight at 55°C. Next day, 1 ml of saturated NaCl solution was added to the cell lysate and left at room temperature for 3 h. DNA extraction was performed using graded ethanol precipitation. The isolated DNA from EOMA cells was used as oxidized DNA whereas DNA from MAE cells was used as nonoxidized control. DNA (1 μg/well) was transfected to MAE cells (7.5 × 104 cells/well) in a 12-well plate using Lipofectamine LTX PLUS reagent as described previously (9). Forty-eight hours after transfection, cells were collected, and Western blot and immunocytochemistry were performed.
Immunoprecipitation and protein expression.
EOMA cells were seeded in a 12-well plate at a density of 0.1 × 106 cells/well 24 h before treatment. Cells were transfected as described above with control and dicer siRNA for 72 h, then washed with ice-cold phosphate-buffered saline (pH 7.4) and lysed with lysis buffer (150 mM KCl, 25 mM Tris-HCl, 5 mM EDTA, 0.5% IgePal, 1 mM PMSF, 1 × protease inhibitor) as described previously (11). Protein concentration was determined using the BCA protein assay (Pierce Biotechnology). TrueBlot anti Rabbit Ig immunoprecipitation (IP) beads (eBiosciences, San Diego, CA) were prewashed with lysis buffer for 40 min. Cell lysates (500 μg) were incubated with prewashed beads for 1 h at 4°C on a rotisserie shaker (Barnstead/Thermolyne, Dubuque, IA). Five to ten micrograms anti-rabbit c-Jun antibody (Santa Cruz Biotechnology, Dallas, TX) was added with the lysate and incubated overnight in a rotisserie shaker at 4°C. The beads were then washed three times with ice-cold lysis buffer (centrifugation at 2,500 g at 4°C for 5 min) and separated into two portions, one for RNA isolation to identify miRNA target genes and another portion for Western blotting to check for successful IP of c-Jun. Immunoprecipitated complexes were washed three times with lysis buffer (centrifugation at 2,500 g at 4°C for 5 min). For Western blot, samples were subjected to SDS/PAGE after reduction with 1 M DTT as previously described (16).
Statistical methods.
All experiments were conducted at least for three samples of each group for two independent replicates unless it is mentioned in the figure legend. Two-sided, two-sample t-test with unequal variance was used to compare the difference between two groups. The residuals from every fitted model were investigated to ensure the assumptions of model were satisfied. Sensitivity analyses were also conducted using nonparametric procedures to ensure the conclusions were robust to the selection of the statistical methods. A P value of <0.05 was considered statistically significant.
RESULTS
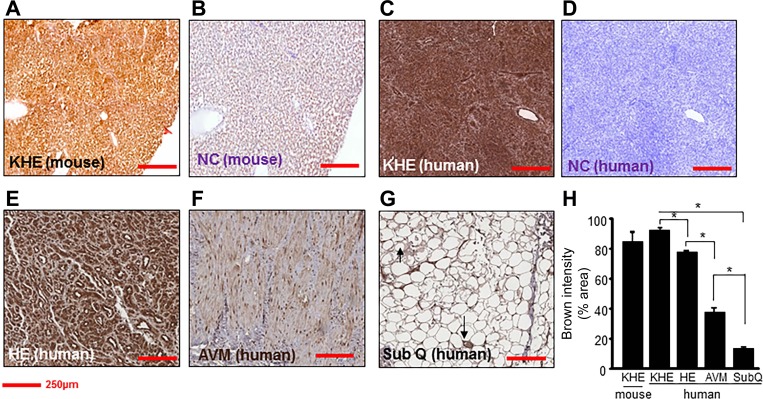
Murine and human EC tumors were stained with Nox-4 antibody to determine relative expression levels compared with non-tumor-forming ECs (Fig. 1, A–G). All human tissue sections were from paraffin-embedded archival tissue specimens, and human hemangioma tumor sections were obtained from children < 5 mo of age to ensure that these tumors were in an actively proliferating state. EOMA cells used in the mouse model generate kaposiform hemangioendotheliomas (KHE), classified as an intermediate grade malignancy, while hemangiomas (HE) are classified as benign tumors. Pixel densitometry of stained sections provided quantitative data as the basis for comparison (Fig. 1H). Nox-4 is normally found in ECs and not surprisingly it was observed in blood vessels in normal human fat albeit at relatively low levels. Arteriovenous malformation (AVM) was included in the analysis as it is also a pathological collection of EC-lined blood vessels. It reflects a high level of EC density, but the Nox-4 expression levels remain relatively low. These results indicate that highly elevated levels of Nox-4 are common to both tumors. Interestingly, the level of Nox-4 expression appears to correlate with the malignant potential of the ECs as it was significantly elevated in HE, with even higher levels of expression observed in KHE. The high levels of expression of Nox-4 identified in both the murine and the human EC tumor sections compared with non-tumor-forming ECs validate the EOMA model used for these experiments as a surrogate for human hemangiomas and indicate the suitability of using this model to test new therapies.
Fig. 1.
Increased expression of Nox-4 in tumor-forming endothelial cells (ECs) compared with non-tumor-forming ECs in humans. Nox-4 immunohistochemistry (1:50) was performed on human and mouse tissue sections. A: EOMA-derived murine kaposiform hemangioendothelioma (KHE). B: secondary antibody alone (NC, mouse). C: human KHE. D: secondary antibody alone (NC, human). E: human hemangioma (HE). F: human arteriovenous malformation (AVM). G: normal ECs in human subcutaneous tissues (SubQ). H: quantification of Nox-4 expression (% area) showed significantly elevated levels of nox-4 protein in tumor-forming ECs compared with non-tumor-forming ECs. Scale bar = 250 μM. Results are expressed as means ± SD of at least 3 determinant sections. *P < 0.05 (n = 3).
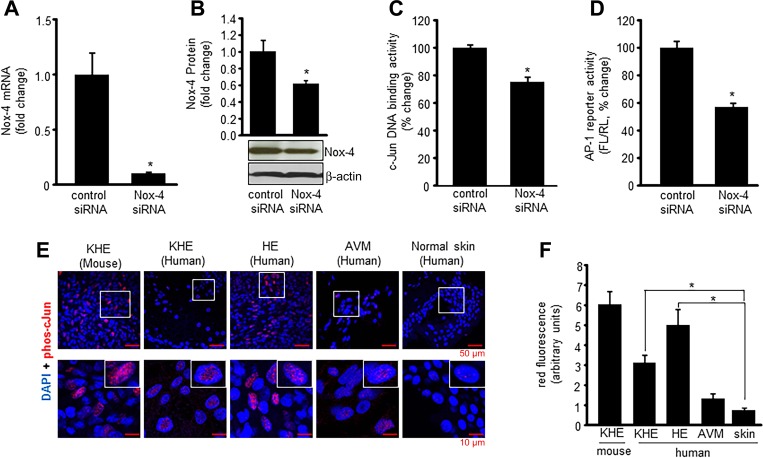
Nox-4 knockdown studies were performed to test the significance of this oxidant-generating enzyme on AP-1 activity (Fig. 2, A and B). An ELISA-based assay (AP-1 family Trans-AMkit, Active Motif) that uses plates coated with specific DNA consensus binding sequences for all potential AP-1 protein subunits (c-Jun, c-Fos, JunD, JunB, FosB, Fra-1) and quantifies binding using a fluorescent antibody that specifically detects the phosphorylated/activated (phospho) form of the protein was used to determine that phospho c-Jun was the predominant AP-1 subunit protein present in the nuclear protein pellet of EOMA cells (data not shown). This same method, using a c-Jun-specific kit, was used on EOMA cells after nox-4 knockdown with siRNA, and there was a significant decrease in nuclear phospho c-Jun DNA binding detected following Nox-4 knockdown compared with siRNA-transfected controls (Fig. 2C). To verify loss of AP-1 transcriptional activity after Nox-4 knockdown, pAP1-Luc reporter plasmids were cotransfected with pRL-cmv Renilla firefly luciferase plasmids into EOMA cells previously treated with either scrambled or nox-4 siRNA. Nox4 knockdown significantly inhibited AP-1 transcriptional activity (Fig. 2D). Immunohistochemical localization of phospho c-Jun in both human and mouse tumor sections (Fig. 2E) detected significantly elevated levels of phospho c-Jun in the nucleus of tumor-forming ECs compared with non-tumor-forming ECs establishing a direct relationship between Nox-4 and c-Jun levels in tumor-forming ECs (Fig. 2F). Collectively these results indicate AP-1 transcriptional activity in tumor-forming ECs is Nox-4 dependent.
Fig. 2.
Nox-4 induces nuclear c-Jun DNA binding and AP-1 transcriptional activation. Successful knockdown of nox-4 after siRNA transfection was confirmed by real-time PCR (mRNA) (A) and Western blot (protein) (B). DNA binding activity (C) of c-Jun from EOMA cell nuclear protein extracts was performed using an ELISA-based (Trans-AM) method. There was significant decrease in c-Jun DNA binding activity with Nox-4 knockdown. D: EOMA cells were transfected with Nox-4 siRNA; after 72-h transfection, cells were washed and cotransfected with a pAP-1 luciferase and pRL-TK Renilla luciferase plasmids for 48 h. Firefly luciferase (FL) activity levels were normalized with Renilla luciferase(RL). Nox-4 knockdown in EOMA cells resulted in loss of AP-1 transcriptional activation. E: confocal images of immunofluorescence detection of phospho-c-Jun in murine and human EC tumor sections with human AVM and normal blood vessel in human skin as EC controls. F: quantification of fluorescence signal intensity using Olympus FV10-ASW software. Results are expressed as means ± SD. *P < 0.05; for tumor sections, n = 5 fields per section.
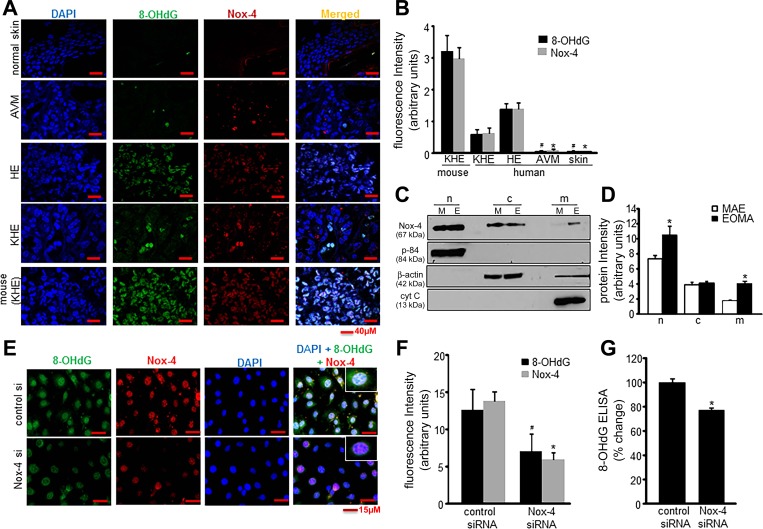
Nox-4 activity has been previously shown to result in elevated levels of 8-OHdG in the urine of mice with KHE tumors from EOMA; thus it was hypothesized that Nox-4 oxidation of DNA was the stimulus for apex-1 expression necessary for AP-1 transcriptional activity. Immunocytochemical staining of murine and human EC tumors was done to determine the relative abundance of 8-OHdG, and it was present in significantly higher levels in tumor-forming ECs (HE and KHE) compared with non-tumor-forming ECs. The amount of 8-OHdG was below detection limits in ECs of normal skin and arteriovenous malformations (Fig. 3, A and B). Since oxidized DNA can also be generated outside the nucleus, cell fractionation was done and Nox-4 levels measured, detected by Western blot for nuclear, cytosol, and mitochondrial membrane fractions. Murine aortic endothelial (MAE) cells, which is a non-tumor-forming cell line, was used as a control. Membrane fractionation studies showed that Nox-4 protein levels were significantly higher in the nuclear fraction compared with any other subcellular fraction and that levels of Nox-4 were higher for all membrane fractions in EOMA compared with MAE cells (Fig. 3, C and D). Immunocytochemistry was done in EOMA cells to demonstrate the relationship between nuclear DNA oxidation and Nox-4. Colocalization of nuclear DAPI staining and 8-OHdG showed a significant decrease in fluorescence intensity in cells treated with nox-4 siRNA compared with control siRNA (Fig. 3, E and F). These imaging observations were validated by ELISA detection of 8-OHdG (Fig. 3G) and confirmed the hypothesis that Nox-4 activity generates oxidized DNA in the nucleus of tumor-forming ECs.
Fig. 3.
Nox-4-derived oxidants induce nuclear DNA oxidation. A: confocal images of immunofluorescence detection of 8-OHdG (1:75) and Nox-4 (1:100) in human tissue sections. DAPI was used to counterstain the nucleus. B: fluorescence intensity analysis (Olympus FV10-ASW software) showed significant differences in both 8-OHdG and Nox-4 detection between tumor-forming and non-tumor-forming ECs; *P < 0.05 for Nox-4 (HE vs. others) and #P < 0.05 for 8-OHdG (HE vs. others) of 5 determinant sections. C: Nox-4 localization in subcellular fractions of nucleus (n), cytosol (c), and mitochondrial membrane (m) was detected by Western blots in both cell types of EOMA (E) and murine aortic endothelial (MAE) (M), which was normalized against housekeeping proteins as described in materials and methods. D: representative band intensity was measured using ImageJ software. E: immunocytochemistry shows colocalization of 8-OHdG (1:100), Nox-4 (1:100), and DAPI (1:10,000) in Nox-4 knockdown EOMA cells. F: fluorescence intensity quantified using Olympus FV10-ASW software shows elevated levels of 8-OHdG and Nox-4 in tumor-forming vs. non-tumor-forming ECs. G: significant decrease in 8-OHdG levels measured by ELISA in EOMA cells after nox-4 siRNA transfection. Results are expressed as means ± SD; *P < 0.05. For tumor sections, n = 5 fields per section.
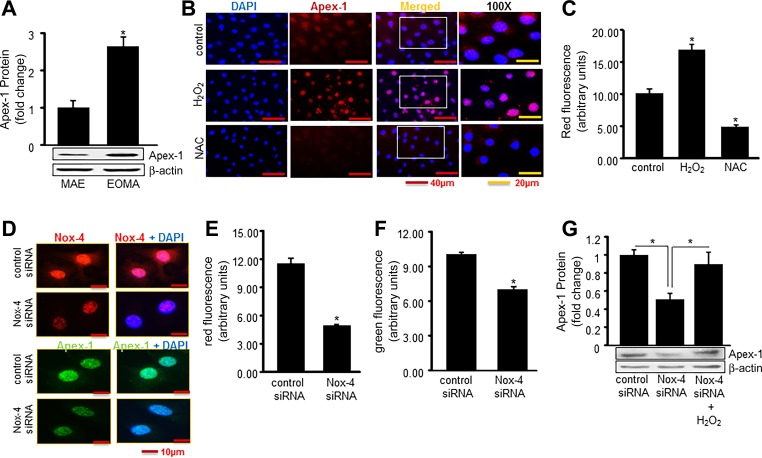
Experiments were then performed to determine whether Nox-4 activity could stimulate Apex-1 expression. Apex-1 protein expression was threefold higher in tumor-forming EOMA cells compared with non-tumor-forming MAE cells, indicating that expression levels are pathologically elevated in tumor-forming ECs (Fig. 4A). To test the involvement of oxidants in Apex-1 expression, EOMA cells were treated with the antioxidant N-acetylcysteine (NAC) for 12 h. The direct involvement of H2O2 was established by demonstrating that Apex-1 is induced by 0.25 mM H2O2 (Figs. 4, B and C). Nox-4 knockdown studies were used to verify that it was the source of oxidant responsible for inducing Apex-1 expression (Fig. 4, D–F). The observation that direct H2O2 treatment could restore induction of Apex-1 in nox-4 knockdown cells established that the oxidant generation component of Nox-4 function was responsible for Apex-1 induction (Fig. 4G).
Fig. 4.
Nox-4-derived H2O2 induces expression of Apex-1. A: Western blot shows elevated Apex-1 protein expression in tumor-forming EOMA cells vs. non-tumor-forming MAE cells. B: H2O2-induced Apex-1 expression was measured by immunocytochemistry. Cells were challenged with H2O2 (250 μM) for 6 h and then treated with N-acetyl cysteine (NAC, 5 mM) for 12 h. C: quantification of fluorescence intensity for Apex-1 was analyzed. D: Apex-1 expression measured by immunocytochemistry was significantly reduced in Nox-4 knockdown cells. E and F: quantification of fluorescence intensity of both Nox-4 and Apex-1. G: rescue effect has been established by posttreatment of H2O2 after knocking down Nox-4. Results are expressed as means ± SD; *P < 0.05.
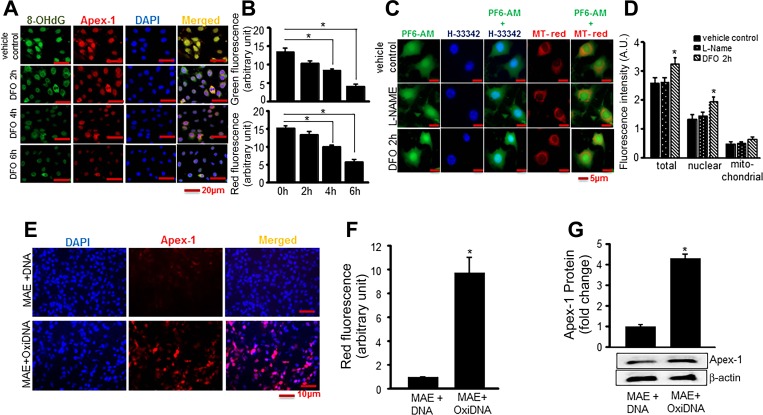
DNA oxidation is not caused directly by H2O2, so the proposed mechanism for oxidation was production of highly reactive hydroxyl radical (HO·) derived from H2O2 and transition metal ions such as Fe2+ that facilitate H2O2 → HO·conversion through Fenton chemistry. To determine whether H2O2-induced DNA damage and Apex-1 expression were Fe2+ dependent, immunofluorescence detection of 8-OHdG and Apex-1 was performed in EOMA cells in the presence or absence of the iron chelator deferoxamine (DFO). There was a progressive decrease in detectable 8-OHdG and Apex-1 with increasing time of exposure to DFO (Fig. 5, A and B). These results were verified by another approach using PF6-AM, a boronate-based fluorescent probe that is irreversibly converted to a brighter fluorescent species in a ratiometric response upon reaction with H2O2. The use of DFO in PF6-AM-treated EOMA cells resulted in accumulation of H2O2 and increasing green fluorescence intensity as H2O2 was spared from being converted to HO·. Since PF6-AM can also react with peroxynitrite, l-NAME was used as a control and the difference in green fluorescence intensity between the control and l-NAME treated cells indicates that the contribution of peroxynitrite was minimal (Fig. 5, C and D). These lines of evidence pointed toward oxidized DNA as a trigger for the induced expression of Apex-1. This hypothesis was validated by experiments where oxidized DNA derived from EOMA cells was added to healthy MAE cells. Oxidized DNA, but not native DNA, induced Apex-1 in MAE cells establishing oxidized DNA as a direct trigger to induce Apex-1 in EOMA cells (Fig. 5, E–G).
Fig. 5.
Nox-4-derived oxidants cause DNA oxidation that stimulates Apex-1 expression. A: accumulation of 8-OHdG in nuclei of EOMA cells was decreased in response to deferoxamine (DFO) treatment. EOMA cells were grown on coverslips. After 18 h, cells were treated with DFO (100 μM) for specified time points as indicated. Cells were fixed and stained with 8-OHdG (green), Apex-1 (red), and DAPI (blue). B: fluorescence intensity determines there was significant decrease of both Apex-1 and 8-OHdG expression with increasing time points. C: accumulation of H2O2 in presence of DFO was determined by a specific cell-permeable dye PF6-AM that interacts with intracellular H2O2 and generates a signal (green). Levels of colocalization were analyzed by nuclear (Hoechst 33342) and mitochondrial (MitoTracker Red) staining. l-NAME was used as inhibitor for peroxynitrite to confirm the specificity of PF6-AM. D: quantification of fluorescence intensity determines increased accumulation of H2O2 after 2 h of DFO treatment in total as well as nuclear compartments. E: images display oxidized DNA increased expression of Apex-1. Oxidative DNA was isolated from EOMA cells and transfected to MAE cells for 48 h as described in Methods. F: quantification of fluorescence intensity of Apex-1 in presence of oxidized DNA. G: Apex-1 protein expression in presence of oxidative DNA measured by Western blot analysis. Results are expressed as means ± SD; *P <0.05.
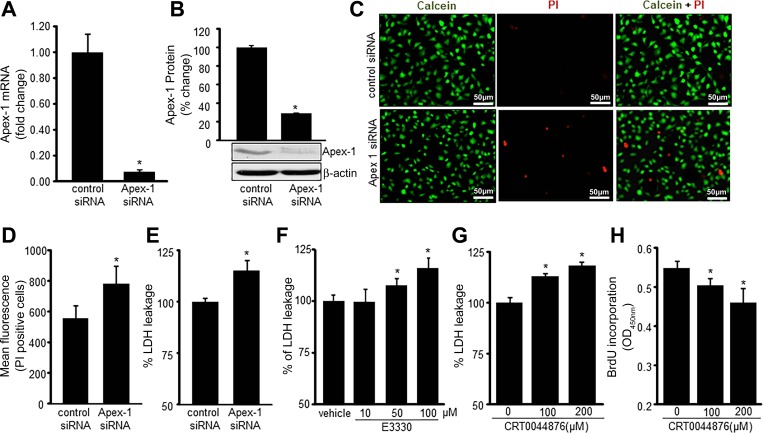
An abundance of nuclear oxidant is known to be lethal for cells. What rescues the EOMA cells from this lethal fate is an overabundance of Apex-1. In EOMA, successful knockdown of Apex-1 (Fig. 6, A and B) resulted in loss of cell viability establishing Apex-1 as a key survival factor in these diseased cells (Fig. 6, C–E). Considering that Apex-1 has two functions, experiments were performed to determine the contribution of these mechanisms to EOMA cell survival. EOMA cells were treated with E3330, a specific inhibitor for Apex-1 redox stabilization function (23), and CRT0044876, a specific inhibitor for Apex-1 base excision repair function (15). Inhibition of each function of Apex-1 resulted in loss of cell viability, establishing that both functions of Apex-1 in EOMA serve as a survival factor in these cells (Fig. 6, F–H).
Fig. 6.
Apex-1 is necessary for EOMA cell survival. Transient transfection with Apex-1 siRNA was analyzed by decreased Apex-1 mRNA (A) and protein (B). C: cell viability was measured by calcein-AM (green, live cells) and propidium iodide (PI; red, dead cells) staining in Apex-1 siRNA transfected cells. D and E: cell viability was also measured by flow cytometry and lactate dehydrogenase (LDH) assay in Apex-1 knockdown cells. F and G: treatment with E3330, an inhibitor for redox function of Apex-1, and CRT0044876, an inhibitor for DNA repair function of Apex-1, showed increased cytotoxicity of EOMA cells as measured using LDH leakage. H: cellular proliferation was measured using BrdU incorporation assay in CRT0044876-treated EOMA cells for 24 h. Results are expressed as means ± SD; *P < 0.05.
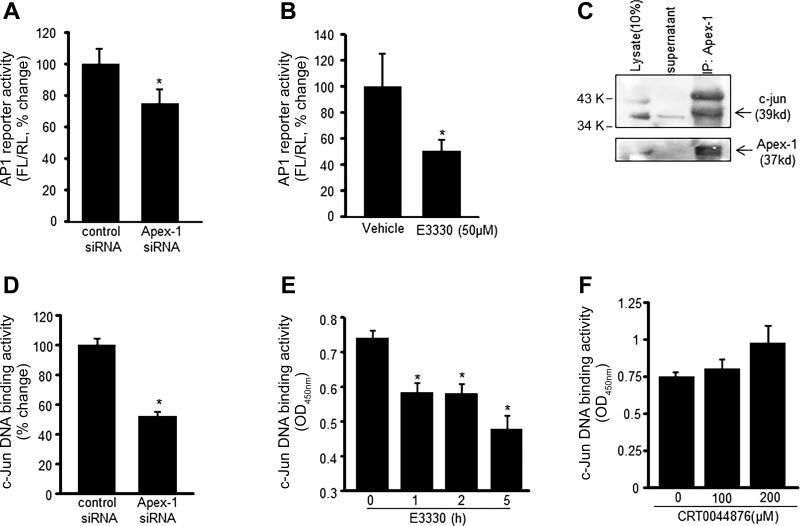
As integral components of EOMA tumor promotion, cell survival must be followed by proliferation. AP-1 transactivation is a key driver of this proliferation process. To determine whether AP-1 transactivation was dependent on the presence of Apex-1, EOMA cells were transfected with either apex-1 siRNA or control siRNA and subsequently cotransfected with a firefly luciferase reporter plasmid under AP-1 control and a renilla luciferase reporter plasmid. AP-1 transactivation was blunted in cells subjected to Apex-1 knockdown demonstrating dependence of the transactivation on Apex-1 (Fig. 7A). Given that transcription factors are known to require a reducing microenvironment to bind to their cognate DNA sites it was hypothesized that the redox stabilization function of Apex-1 accounted for its ability to support AP-1 transactivation. EOMA cells cotransfected with AP-1 firefly and renilla luciferase reporter vectors were treated with E3330, which selectively targets Apex-1 redox activity. The E3330-treated cells had a significant decrease in AP-1 transactivation to provide specific evidence that the redox stabilization function of Apex-1 is necessary for AP-1 transactivation (Fig. 7B). Physical interaction between Apex-1 and AP-1 was demonstrated by coimmunoprecipitation (Fig. 7C). Additionally, decreased nuclear c-Jun DNA binding after apex-1 knockdown confirmed the critical significance of Apex-1 in AP-1 DNA binding (Fig. 7D). EOMA cells treated with E3330, but not CRT044876 (Fig. 7F), showed time-dependent decrease in nuclear c-Jun DNA binding (Fig. 7E).
Fig. 7.
Apex-1 promotes AP-1 activation and c-Jun DNA binding. A and B: AP-1 transcription activity was analyzed in EOMA cells. A: cells were transfected with control and Apex-1 siRNA; after 72-h transfection, cells were washed and cotransfected with a pAP-1 luciferase and pRL-TK Renilla luciferase plasmids for 48 h. Firefly luciferase activity levels were normalized with Renilla luciferase. B: AP-1 activity was measured in E3330 (50 μM, 5 h)-treated EOMA cells. C: association of c-Jun with Apex-1 was identified by coimmunoprecipitation (IP). The cell lysate were subjected to immunoprecipitation with Apex-1 antibody. The immunoprecipitates were subjected to SDS PAGE and immunoblotting (IB) for c-Jun and Apex-1. D and E: c-Jun DNA binding ability was measured in Apex-1 knockdown (D) and also in E3330 (50 μM, E)-treated EOMA cells using an ELISA-based (Trans-AM) method. F: treatment of CRT0044876 did not show any significant effects on c-Jun DNA binding activity. Results are expressed as means ± SD; *P < 0.05.
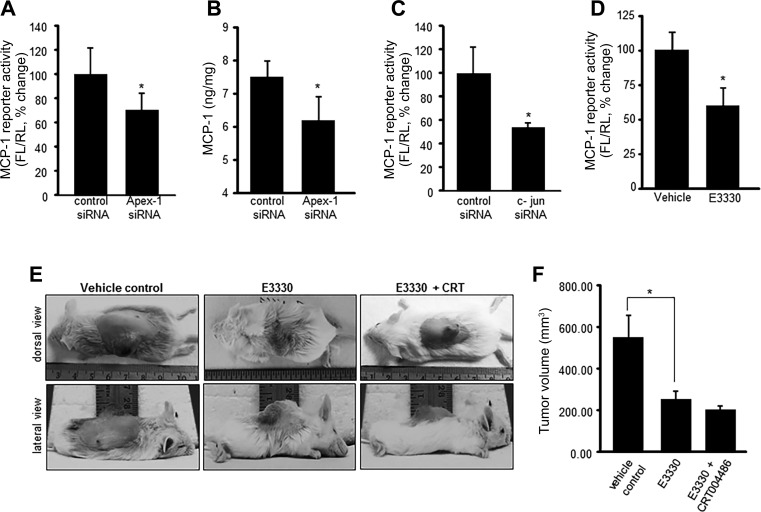
EOMA derived EC tumor progression depends on cell proliferation and MCP-1 expression. AP-1 serves as a critical hub as it supports both of these processes. Apex-1 knockdown, which we now know blunts AP-1 activation (Fig. 7), downregulated MCP-1 expression (Fig. 8, A and B). Comparable blunting of MCP-1 expression was achieved by specific knockdown of the AP-1 protein c-Jun (Fig. 8C). The observation that results of Apex-1 knockdown on MCP-1 transactivation could be reproduced by treatment of cells with E3330 establishes that it was the redox factor function of Apex-1 that was responsible for induced MCP-1 expression (Fig. 8D).
Fig. 8.
Redox function of Apex-1 is required for MCP-1 activation and EC tumor growth in vivo. Apex-1 knockdown in EOMA cells resulted in significant decrease in MCP-1 reporter activity (A), and MCP-1 release in the media was measured by ELISA (B). C: MCP-1 reporter activity was significantly decreased in c-Jun knockdown EOMA cells and in E3330 (50 μM, 5 h)-treated cells (D). Redox changes of Apex-1 influences HE outcome in vivo. E: tumor growth rates were evaluated after 7 days of E3330 treatment (25 mg/kg ip twice daily) alone and in combination with CRT0044876 (10 mg/kg ip twice daily). F: tumor volume was quantified using calipers (length × width × height). Results are expressed as means ± SD; *P < 0.05.
To determine the clinical significance of Apex-1 function to EC tumor formation, KHE were generated in 129P3 mice by subcutaneous injection with EOMA cells. Intraperitoneal injection of E3330 or E3330 + CRT0044876 was started on day 0. On day 7 mice were euthanized because that is when they start to die from the tumor. E3330 treatment resulted in 50% decrease in tumor size, but the addition of CRT0044876 to E330 resulted in minimal additional tumor growth inhibition (Fig. 8, E and F). These results indicate that the tumor growth is more dependent on the redox functions of Apex-1 and uphold targeting the redox function of Apex-1 and modification of the oxidative environment as valid therapeutic strategies for the treatment of EC tumors.
DISCUSSION
Nox-4 is known to be constitutively expressed in ECs (37), but these are the first reported results demonstrating elevated levels of Nox-4 protein expression in tumor-forming compared with non-tumor-forming ECs in humans. Elevated levels of Nox-4 have been previously reported for several other tumors including breast, thyroid, glioblastoma, and melanoma (8, 35, 51, 59). The fact that EC tumors share a common pathological finding of elevated Nox-4 activity with other tumor types suggests that this source of ROS production and oxidative stress plays a fundamental role in malignant transformation. It also indicates that therapeutic strategies that target this pathway may have utility for other tumor types, highlighting the clinical significance of this work.
There have been reports that AP-1 can induce transcriptional activation of Nox-4 (2, 40), which suggests there is a positive feedback loop between Nox-4 and AP-1. This could explain the strikingly high levels of Nox-4 and c-Jun observed in tumor-forming ECs. Our results are consistent with the observations that constitutive expression of AP-1 is associated with tumor promotion and aberrant cellular proliferation (46, 47). ROS have been recognized as promoters of carcinogenesis (6, 29). These results demonstrate that nox-4 derived H2O2 and OH·are produced in excess in tumor-forming ECs. An undesirable consequence of ROS production in tumor-forming ECs is activation of oxidant-sensitive transcription factors such as AP-1 and induction of chemokines like MCP-1 that stimulate angiogenesis and promote tumor growth. Tumors of EC origin are unique because angiogenesis is the sole mechanism for tumor growth (52). EOMA cells are completely dependent on Nox-4-induced AP-1 activation and expression of MCP-1 to drive angiogenesis and tumor growth (13, 17). It is not merely a supportive process as it is for other solid tumors. ROS-stimulated angiogenesis has been shown to be Apex-1 dependent (31, 54). Thus EC tumors may be more reliant on Apex-1 expression as a survival mechanism than other tumors.
Several lines of evidence implicate Apex-1 in cancer cell growth and tumorigenicity. Apex-1 plays a role in sustaining cell viability and proliferation in colon cancer and breast cancer cells (11). Depletion of Apex-1 significantly suppressed tumor promoter-induced colony formation of mouse epithelial cells (60), and inhibition of the Apex-1 redox domain suppressed tumor-associated EC growth and angiogenesis (61). Elevated Apex-1 protein levels have been found in many different tumors and correlate inversely with prognosis (30, 42), indicating that elevated Apex-1 levels are potentially clinically significant. This work is the first to report that Nox-4-derived ROS induce Apex-1 expression in tumor-forming ECs.
Colocalization of Nox-4, Apex-1 and oxidative events to the nucleus provide important context for interpreting these results. Introduction of ROS into the nucleus is usually lethal to cells, but EOMA cells are able to withstand this challenge due to the induction of Apex-1. Although oxidatively damaged DNA and AP-1 have been shown to induce Apex-1 expression (5, 34, 45, 57), these are the first reported results to demonstrate that Nox-4-derived oxidants induce Apex-1 expression, specifically through Fe2+-dependent conversion of H2O2 to hydroxyl radical, resulting in elevated levels of 8-OHdG in tumor-forming ECs. Elevated levels of 8-OHdG and Apex-1 have been observed clinically in many tumors and correlate positively with malignant behavior (12, 50), suggesting that oxidative modification of DNA and induction of Apex-1 expression may be central biological events occurring in many different types of tumors in addition to EC tumors.
The positive feedback mechanisms between Nox-4, AP-1, and subsequent induction of Apex-1 expression links these three proteins in an important pathological mechanism responsible for promoting EC tumor growth. Apex-1 represents the foundation for EC tumor growth because without it AP-1 could not bind to the MCP-1 promoter to stimulate angiogenesis and cells would not survive the oxidative DNA damage induced by Nox-4. These are also the first reported results to demonstrate that EC tumor formation is Apex-1 dependent. Thus Apex-1 is the critical survival factor for these cells and without it there would be a cascade of events leading to loss of proliferation and cell death. Collectively, these results support the claim that Apex-1 is a viable therapeutic target for EC tumors.
GRANTS
This work was supported by National Institute of General Medical Sciences Grant R01-GM-095657 (G. M. Gordillo).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.B., S.K., C.K.S., and G.M.G. conception and design of research; A.b. performed experiments; A.B., S.K., S.R., X.P.P., C.K.S., and G.M.G. analyzed data; A.B., S.K., S.R., X.P.P., C.K.S., and G.M.G. interpreted results of experiments; A.b. prepared figures; A.b., S.K., S.R., X.P.P., C.K.S., and G.M.G. drafted manuscript; A.B., S.K., S.R., X.P.P., C.K.S., and G.M.G. edited and revised manuscript; A.B., S.K., S.R., X.P.P., C.K.S., and G.M.G. approved final version of manuscript.
REFERENCES
- 1.Atalay M, Gordillo G, Roy S, Rovin B, Bagchi D, Bagchi M, Sen CK. Anti-angiogenic property of edible berry in a model of hemangioma. FEBS Lett 544: 252–257, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Bai G, Hock TD, Logsdon N, Zhou Y, Thannickal VJ. A far-upstream AP-1/Smad binding box regulates human NOX4 promoter activation by transforming growth factor-beta. Gene 540: 62–67, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee J, Das Ghatak P, Roy S, Khanna S, Sequin EK, Bellman K, Dickinson BC, Suri P, Subramaniam VV, Chang CJ, Sen CK. Improvement of human keratinocyte migration by a redox active bioelectric dressing. PLoS One 9: e89239, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bapat A, Fishel ML, Kelley MR. Going ape as an approach to cancer therapeutics. Antioxid Redox Signal 11: 651–668, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal 11: 621–638, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhat HK, Calaf G, Hei TK, Loya T, Vadgama JV. Critical role of oxidative stress in estrogen-induced carcinogenesis. Proc Natl Acad Sci USA 100: 3913–3918, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boonstra J, Post JA. Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene 337: 1–13, 2004. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho DP, Dupuy C. Role of the NADPH oxidases DUOX and NOX4 in thyroid oxidative stress. Eur Thyroid J 2: 160–167, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das A, Ganesh K, Khanna S, Sen CK, Roy S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol 192: 1120–1129, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elgharably H, Ganesh K, Dickerson J, Khanna S, Abas M, Ghatak PD, Dixit S, Bergdall V, Roy S, Sen CK. A modified collagen gel dressing promotes angiogenesis in a preclinical swine model of chronic ischemic wounds. Wound Repair Regen 22: 720–729, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell 17: 463–470, 2005. [DOI] [PubMed] [Google Scholar]
- 12.Futagami S, Hiratsuka T, Shindo T, Horie A, Hamamoto T, Suzuki K, Kusunoki M, Miyake K, Gudis K, Crowe SE, Tsukui T, Sakamoto C. Expression of apurinic/apyrimidinic endonuclease-1 (APE-1) in H. pylori-associated gastritis, gastric adenoma, and gastric cancer. Helicobacter 13: 209–218, 2008. [DOI] [PubMed] [Google Scholar]
- 13.Gordillo G, Fang H, Khanna S, Harper J, Phillips G, Sen CK. Oral administration of blueberry inhibits angiogenic tumor growth and enhances survival of mice with endothelial cell neoplasm. Antioxid Redox Signal 11: 47–58, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordillo G, Fang H, Park H, Roy S. Nox-4-dependent nuclear H2O2 drives DNA oxidation resulting in 8-OHdG as urinary biomarker and hemangioendothelioma formation. Antioxid Redox Signal 12: 933–943, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordillo GM, Atalay M, Roy S, Sen CK. Hemangioma model for in vivo angiogenesis: inducible oxidative stress and MCP-1 expression in EOMA cells. Methods Enzymol 352: 422–432, 2002. [DOI] [PubMed] [Google Scholar]
- 16.Gordillo GM, Biswas A, Khanna S, Pan X, Sinha M, Roy S, Sen CK. Dicer knockdown inhibits endothelial cell tumor growth via microRNA 21a-3p targeting of Nox-4. J Biol Chem 289: 9027–9038, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordillo GM, Onat D, Stockinger M, Roy S, Atalay M, Beck FM, Sen CK. A key angiogenic role of monocyte chemoattractant protein-1 in hemangioendothelioma proliferation. Am J Physiol Cell Physiol 287: C866–C873, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Grimm S, Jennek S, Singh R, Enkelmann A, Junker K, Rippaus N, Berndt A, Friedrich K. Malignancy of bladder cancer cells is enhanced by tumor-associated fibroblasts through a multifaceted cytokine-chemokine loop. Exp Cell Res 335: 1–11, 2015. [DOI] [PubMed] [Google Scholar]
- 19.Hacke K, Rincon-Orozco B, Buchwalter G, Siehler SY, Wasylyk B, Wiesmuller L, Rosl F. Regulation of MCP-1 chemokine transcription by p53. Mol Cancer 9: 82, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haggstrom AN, Drolet BA, Baselga E, Chamlin SL, Garzon MC, Horii KA, Lucky AW, Mancini AJ, Metry DW, Newell B, Nopper AJ, Frieden IJ. Prospective study of infantile hemangiomas: clinical characteristics predicting complications and treatment. Pediatrics 118: 882–887, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Hoak JC, Warner ED, Cheng HF, Fry GL, Hankenson RR. Hemangioma with thrombocytopenia and microangiopathic anemia (Kasabach-Merritt syndrome): an animal model. J Lab Clin Med 77: 941–950, 1971. [PubMed] [Google Scholar]
- 22.Jacobs AH, Walton RG. The incidence of birthmarks in the neonate. Pediatrics 58: 218–222, 1976. [PubMed] [Google Scholar]
- 23.Kelley MR, Luo M, Reed A, Su D, Delaplane S, Borch RF, Nyland RL 2nd, Gross ML, Georgiadis MM. Functional analysis of novel analogues of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref-1. Antioxid Redox Signal 14: 1387–1401, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khanna S, Park HA, Sen CK, Golakoti T, Sengupta K, Venkateswarlu S, Roy S. Neuroprotective and antiinflammatory properties of a novel demethylated curcuminoid. Antioxid Redox Signal 11: 449–468, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khanna S, Roy S, Ryu H, Bahadduri P, Swaan PW, Ratan RR, Sen CK. Molecular basis of vitamin E action: tocotrienol modulates 12-lipoxygenase, a key mediator of glutamate-induced neurodegeneration. J Biol Chem 278: 43508–43515, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khanna S, Roy S, Slivka A, Craft TK, Chaki S, Rink C, Notestine MA, DeVries AC, Parinandi NL, Sen CK. Neuroprotective properties of the natural vitamin E alpha-tocotrienol. Stroke 36: 2258–2264, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HW, Murakami A, Williams MV, Ohigashi H. Mutagenicity of reactive oxygen and nitrogen species as detected by co-culture of activated inflammatory leukocytes and AS52 cells. Carcinogenesis 24: 235–241, 2003. [DOI] [PubMed] [Google Scholar]
- 28.Kim JS, Diebold BA, Babior BM, Knaus UG, Bokoch GM. Regulation of Nox1 activity via protein kinase A-mediated phosphorylation of NoxA1 and 14-3-3 binding. J Biol Chem 282: 34787–34800, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Klaunig JE, Kamendulis LM, Hocevar BA. Oxidative stress and oxidative damage in carcinogenesis. Toxicol Pathol 38: 96–109, 2010. [DOI] [PubMed] [Google Scholar]
- 30.Koukourakis MI, Giatromanolaki A, Kakolyris S, Sivridis E, Georgoulias V, Funtzilas G, Hickson ID, Gatter KC, Harris AL. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int J Radiat Oncol Biol Phys 50: 27–36, 2001. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Liu X, Zhou T, Kelley MR, Edwards P, Gao H, Qiao X. Inhibition of APE1/Ref-1 redox activity rescues human retinal pigment epithelial cells from oxidative stress and reduces choroidal neovascularization. Redox Biol 2: 485–494, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin VS, Dickinson BC, Chang CJ. Boronate-based fluorescent probes: imaging hydrogen peroxide in living systems. Methods Enzymol 526: 19–43, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo M, Delaplane S, Jiang A, Reed A, He Y, Fishel M, Nyland RL 2nd Borch RF, Qiao X, Georgiadis MM, Kelley MR. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule inhibition of the redox function of Ape1. Antioxid Redox Signal 10: 1853–1867, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merluzzi S, Moretti M, Altamura S, Zwollo P, Sigvardsson M, Vitale G, Pucillo C. CD40 stimulation induces Pax5/BSAP and EBF activation through a APE/Ref-1-dependent redox mechanism. J Biol Chem 279: 1777–1786, 2004. [DOI] [PubMed] [Google Scholar]
- 35.Mondol AS, Tonks NK, Kamata T. Nox4 redox regulation of PTP1B contributes to the proliferation and migration of glioblastoma cells by modulating tyrosine phosphorylation of coronin-1C. Free Radic Biol Med 67: 285–291, 2014. [DOI] [PubMed] [Google Scholar]
- 36.Mulliken JB. Diagnosis and natural history of hemangiomas. In: Vascular Birthmarks: Hemangiomas and Malformations, edited by Mulliken JB, Young AE. Philadelphia, PA: Saunders, 1988, p. 41–62. [Google Scholar]
- 37.Muzaffar S, Jeremy JY, Angelini GD, Shukla N. NADPH oxidase 4 mediates upregulation of type 4 phosphodiesterases in human endothelial cells. J Cell Physiol 227: 1941–1950, 2012. [DOI] [PubMed] [Google Scholar]
- 38.Ohsaki Y, O'Connor P, Mori T, Ryan RP, Dickinson BC, Chang CJ, Lu Y, Ito S, Cowley AW Jr. Increase of sodium delivery stimulates the mitochondrial respiratory chain H2O2 production in rat renal medullary thick ascending limb. Am J Physiol Renal Physiol 302: F95–F102, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park HA, Khanna S, Rink C, Gnyawali S, Roy S, Sen CK. Glutathione disulfide induces neural cell death via a 12-lipoxygenase pathway. Cell Death Differ 16: 1167–1179, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedruzzi E, Guichard C, Ollivier V, Driss F, Fay M, Prunet C, Marie JC, Pouzet C, Samadi M, Elbim C, O'Dowd Y, Bens M, Vandewalle A, Gougerot-Pocidalo MA, Lizard G, Ogier-Denis E. NAD(P)H oxidase Nox-4 mediates 7-ketocholesterol-induced endoplasmic reticulum stress and apoptosis in human aortic smooth muscle cells. Mol Cell Biol 24: 10703–10717, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piechaczyk M, Farras R. Regulation and function of JunB in cell proliferation. Biochem Soc Trans 36: 864–867, 2008. [DOI] [PubMed] [Google Scholar]
- 42.Puglisi F, Aprile G, Minisini AM, Barbone F, Cataldi P, Tell G, Kelley MR, Damante G, Beltrami CA, Di Loreto C. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res 21: 4041–4049, 2001. [PubMed] [Google Scholar]
- 43.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Prot 1: 1458–1461, 2006. [DOI] [PubMed] [Google Scholar]
- 44.Sen CK, Packer L. Antioxidant and redox regulation of gene transcription. FASEB J 10: 709–720, 1996. [DOI] [PubMed] [Google Scholar]
- 45.Sengupta S, Mitra S, Bhakat KK. Dual regulatory roles of human AP-endonuclease (APE1/Ref-1) in CDKN1A/p21 expression. PLoS One 8: e68467, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaulian E. AP-1–The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal 22: 894–899, 2010. [DOI] [PubMed] [Google Scholar]
- 47.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene 20: 2390–2400, 2001. [DOI] [PubMed] [Google Scholar]
- 48.Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal 8: 243–270, 2006. [DOI] [PubMed] [Google Scholar]
- 49.Su D, Delaplane S, Luo M, Rempel DL, Vu B, Kelley MR, Gross ML, Georgiadis MM. Interactions of apurinic/apyrimidinic endonuclease with a redox inhibitor: evidence for an alternate conformation of the enzyme. Biochemistry 50: 82–92, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sumiyoshi S, Kobayashi Y, Kawamura K, Kawata K, Nakamura H. Differential expression of hepatic apurinic/apyrimidinic endonuclease 1, a DNA repair enzyme, in chronic hepatitis. World J Hepatol 5: 206–213, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svensson S, Abrahamsson A, Vazquez Rodriguez G, Olsson AK, Jensen L, Cao Y, Dabrosin C. CCL2 and CCL5 are novel therapeutic targets for estrogen-dependent breast cancer. Clin Cancer Res 2015. April 21 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Mulliken JB, Kozakewich HP, Rogers RA, Folkman J, Ezekowitz RA. Cellular markers that distinguish the phases of hemangioma during infancy and childhood. J Clin Invest 93: 2357–2364, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan LP, Seinen E, Duns G, de Jong D, Sibon OC, Poppema S, Kroesen BJ, Kok K, van den Berg A. A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res 37: e137, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal 11: 601–620, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vesely PW, Staber PB, Hoefler G, Kenner L. Translational regulation mechanisms of AP-1 proteins. Mutat Res 682: 7–12, 2009. [DOI] [PubMed] [Google Scholar]
- 56.Wang C, Quevedo ME, Lannutti BJ, Gordon KB, Guo D, Sun W, Paller AS. In vivo gene therapy with interleukin-12 inhibits primary vascular tumor growth and induces apoptosis in a mouse model. J Invest Dermatol 112: 775–781, 1999. [DOI] [PubMed] [Google Scholar]
- 57.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther 3: 679–686, 2004. [PubMed] [Google Scholar]
- 58.Warner ED, Hoak JC, Fry Hemangioma GL, thrombocytopenia anemia. The Kasabach-Merritt syndrome in an animal model. Arch Pathol 91: 523–528, 1971. [PubMed] [Google Scholar]
- 59.Yamaura M, Mitsushita J, Furuta S, Kiniwa Y, Ashida A, Goto Y, Shang WH, Kubodera M, Kato M, Takata M, Saida T, Kamata T. NADPH oxidase 4 contributes to transformation phenotype of melanoma cells by regulating G2-M cell cycle progression. Cancer Res 69: 2647–2654, 2009. [DOI] [PubMed] [Google Scholar]
- 60.Yang S, Misner BJ, Chiu RJ, Meyskens FL Jr. Redox effector factor-1, combined with reactive oxygen species, plays an important role in the transformation of JB6 cells. Carcinogenesis 28: 2382–2390, 2007. [DOI] [PubMed] [Google Scholar]
- 61.Zou GM, Karikari C, Kabe Y, Handa H, Anders RA, Maitra A. The Ape-1/Ref-1 redox antagonist E3330 inhibits the growth of tumor endothelium and endothelial progenitor cells: therapeutic implications in tumor angiogenesis. J Cell Physiol 219: 209–218, 2009. [DOI] [PubMed] [Google Scholar]