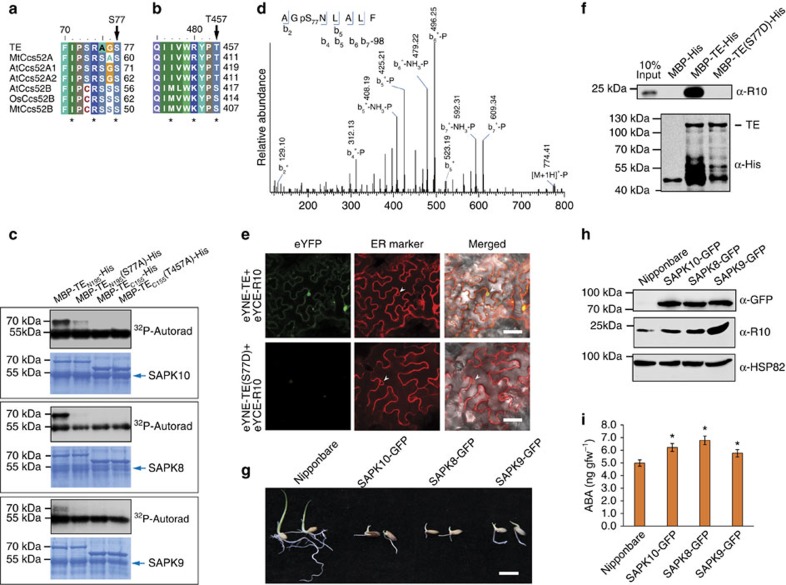
Figure 3. SnRK2s repress the activity of APC/CTE.
(a and b) Analysis of the TE protein sequence identifies two conserved sites, S77 (a) and T457(b), that are potential sites for phosphorylation by SnRK2s. * shows the key amino acids needed for SnRK2 recognition. (c) In vitro phosphorylation assay shows that the TE N-terminus, MBP-TEN195, which contains the S77 site, but not the MBP-TEN195(S77A) mutant, is phosphorylated by all three rice SnRK2s (SAPK8, SAPK9 and SAPK10); whereas neither MBP-TEC155, which contains the T457 site, nor the mutant MBP-TEC155(T457A), are phosphorylated by any of the three SnRK2s. (d) The phosphorylated TE pS77 peptide corresponding to residues 75–82. Observed b-ions and those resulting from the neutral loss of H3PO4 (−98 Da), or both H3PO4 and NH3 (−115 Da) are indicated. (e) BiFC analysis shows the interactions between TE or TE (S77D) and R10. White arrowheads show the nuclear membrane. Scale bar, 100 μm. (f) In vitro pull-down assay shows that MBP-TE-His, but not the mutant MBP-TE(S77D)-His, pulls down R10 (upper panel) from te plant extracts. The lower panel shows that roughly equal amounts of MBP-His, MBP-TE-His and MBP-TE (S77D)-His proteins were used. (g) The SAPK10-GFP, SAPK8-GFP and SAPK9-GFP overexpression lines showed delayed germination and seedling growth compared to the Nipponbare vector control. Scale bar, 1 cm. (h) Western blot analysis shows the levels of R10 protein in Nipponbare (vector control), SAPK10-GFP, SAPK8-GFP and SAPK9-GFP overexpression rice lines. The ‘α-HSP82' signal shows that roughly equal amounts of total plant extract were used. (i) Measurement of ABA levels in Nipponbare (vector control), SAPK10-GFP, SAPK8-GFP and SAPK9-GFP overexpression rice lines. Values are means±s.d. (n=4 replicates). All plants are 10-day-old. Student's t-test analysis indicated a significant difference (compared with Nipponbare, *P<0.05). gfw, gram fresh weight.