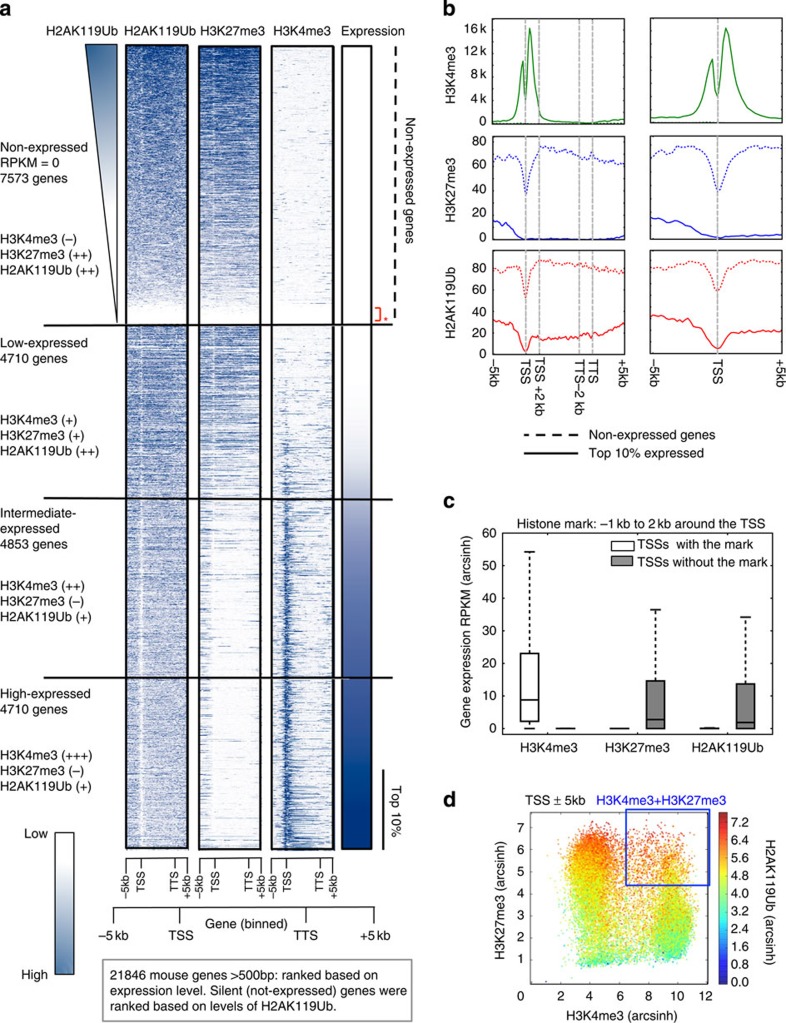
Figure 4. H2AK119Ub marks are present at the expressed and non-expressed genes in EML cells.
Mononucleosomes prepared from EML cells transduced with MiG-empty retrovirus were immunoprecipitated with H2AK119Ub, H3K27me3 and H3K4me3 antibodies, and subjected to ChIP-seq. (a) Genes were ranked based on levels of expression. Shown alongside is the genome-wide distribution of H2AK119Ub, H3K27me3 and H3K4me3 marks across the gene body and ±5 kb around the gene body of all genes >500 bp in length. Based on expression levels in EML cells, we categorized genes into four categories: non-expressed, low, intermediate and high expressed. Non-expressed genes were ranked based on levels of H2AK119Ub. Areas around TSS (−5 kbp, 0 bp) and around TTS (0 bp, +5k bp) were both divided into twenty-five 200-bp bins. Gene bodies were divided into 50 bins of equal width. Genes <500 bp were excluded due to visualization artefacts caused by gene binning. (b) Average input-corrected levels of H2AK119Ub, H3K4me3 and H3K27me3 marks across non-expressed genes and top 10% of genes sorted by expression are represented in two formats, across gene body and ±5 kb around the gene (left) and ±5 kb around TSSs (right). (c) Presence of H2AK119Ub, H3K27me3 and H3K4me3 marks was defined based on ≥10% overlap between SICER peak calls and the area −1 kb to +2 kb around the TSSs. RPKM (reads per kilobase per million) values of genes marked by or lacking the said mark are represented as a box showing the 25th, 50th and 75th percentiles, and whiskers extending to 1.5 times interquartile range. (d) To determine the overlaps between TSSs marked by the H2AK119Ub, H3K4me3 and H3K27me3, enrichments of H3K4me3, H2AK119Ub and H3K27me3 were calculated with respect to input signal in 200 bp bins ±5 kb around each TSS. Total enrichment for each TSS was determined by summing the enrichment values of the ±5 kb area; H2AK119Ub enrichement was colour coded as indicated on the right.