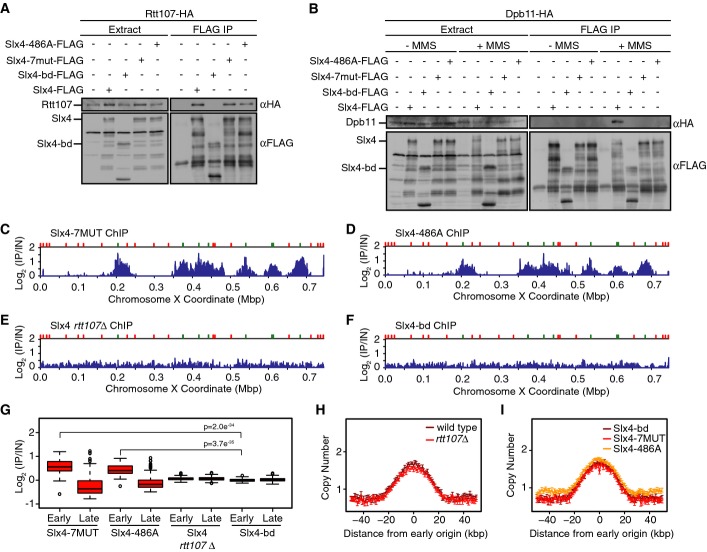
Figure 2.
Slx4 recruitment behind DNA replication forks requires physical interaction with Rtt107
- A, B Analysis of Slx4, Slx4-bd, Slx4-7MUT, and Slx4-486A binding to Rtt107 (A) and Dpb11 (B) by co-immunoprecipitation. The input extract and the anti-Flag immunoprecipitates are shown. Immunoblots were probed with anti-HA or anti-Flag antibodies, as indicated. Binding to Dpb11 was tested with and without treatment of asynchronous cells with 0.035% MMS for 2 h.
- C–F ChIP-seq analysis of Slx4 was performed following synchronous release of (C) slx4-7MUT-FLAG, (D) slx4-486A-FLAG, (E) SLX4-FLAG rtt107Δ, and (F) slx4-bd-FLAG cells into S phase in the presence of 0.035% MMS for 60 min. Slx4 ChIP enrichment scores on chromosome 10 are shown.
- G The distributions of Slx4 ChIP enrichment scores at early- and late-firing origins genome-wide are shown as a boxplot for slx4-7MUT-FLAG, slx4-486A-FLAG, SLX4-FLAG rtt107Δ, and slx4-bd-FLAG cells. The median is indicated by the horizontal bar, the box spans the first through third quartiles, the whiskers extend to the last data points within 1.5 times the interquartile range, and outliers are plotted as circles. The distributions at early origins were compared using the Wilcoxon rank-sum test. The P-values for Slx4-7MUT vs Slx4-bd and Slx4-486A vs Slx4-bd are shown. The P-values for Slx4-7MUT and Slx4-486A vs Slx4 in rtt107Δ were similar, < 10−33.
- H, I The median (± standard error) copy number across n = 108 early-firing origins is plotted for wild-type and rtt107Δ cells (H) and for slx4-7MUT, slx4-486A, and slx4-bd cells (I) following synchronous release into S phase in the presence of 0.035% MMS for 60 min.
Source data are available online for this figure.