Abstract
Identifying proteins that recognize histone methylation is critical for understanding chromatin function. Vermeulen et al. (2010) now describe a cutting-edge strategy to identify and characterize several nuclear proteins and complexes that recognize five major histone trimethyl marks.
In eukaryotes, DNA does not exist in a free state but rather is packaged and compacted by histones and other proteins into the higher order structure of chromatin. One of the major mechanisms for regulating chromatin function involves reversible covalent methylation of specific lysine residues within histone proteins. For example, trimethylation of histone H3 at lysine 4 (H3K4me3) is most frequently found at the promoters of actively transcribed genes, whereas trimethylation of histone H3 at lysine 9 (H3K9me3) is enriched at silent chromatin. The distinct genomic distribution of these and other histone methylation marks has led to the proposal that methylation establishes discrete functional states, raising the question of how the addition of methyl moieties to histones, which does not intrinsically alter chromatin structure, can affect physiologic nuclear programs. It is now understood that specialized chromatin-regulatory factors, named “readers,“ have evolved to specifically recognize distinct histone modifications and that these readers define the functional consequence of histone methylation (Figure 1) (Taverna et al., 2007). In this issue, Vermeulen et al. (2010) integrate multiple technologies to identify readers of five major histone trimethyl marks: at H3K4, H3K9, H3K27, H3K36, and H4K20. The authors determine the genome-wide occupancy of the readers and provide evidence that many of them are present in complexes that regulate gene expression and DNA replication.
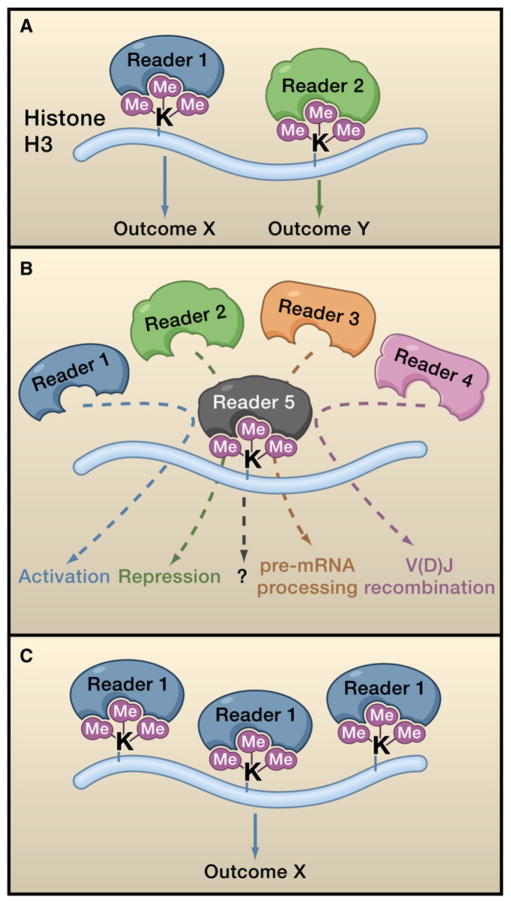
Figure 1. Readers Determine the Effects of Lysine Methylation.
The schematic depicts three different models for how methyllysine readers can sense and transduce histone methylation marks into biological outcomes.
(A) A specific reader recognizes one mark and links it to one specific outcome. In this way, distinct readers link different marks to alternate functional outcomes.
(B) Five distinct readers all specifically recognize the same mark, for example trimethylated histone H3 lysine 4 (H3K4me3), resulting in five alternate biological outputs being linked to the one mark.
(C) One reader binds to multiple different marks (such as H3K9me3, H3K27me3, and H4K20me3), linking the three different marks to a single biological output.
To date, readers with methyllsyine-binding activity have been identified in members of seven different protein domain families: chromo, tudor, PHD finger, MBT, PWWP, Ankryin repeats, and WD40 repeats (Taverna et al., 2007). It is common for proteins within chromatin-regulatory complexes to contain one or more of these methyllsyine-binding motifs and for this binding activity to play an important role in determining the genome-wide distribution of the complex. For example, the basal transcription factor TFIID binds to and is localized at genomic sites enriched with H3K4me3 through a PHD finger present in the TFIID component TAF3 (Vermeulen et al., 2007). The TFIID complex was identified as an H3K4me3-interacting complex in a screen based on SILAC (stable isotope labeling by amino acids in cell culture) proteomics (Vermeulen et al., 2007). The current work by Vermeulen et al. employs an expanded and updated SILAC-based proteomic screen to identify proteins that bind two marks associated with transcription activation (H3K4me3 and H3K36me3) and three marks linked to transcriptional repression (H3K9me3, H3K27me3, and H4K20me3.) Their approach uses quantitative mass spectrometry to identify proteins from nuclear extracts of human cells that preferentially bind to methylated peptides.
Vermeulen and colleagues identify a number of known H3K4me3 readers, for example TAF3 (Vermeulen et al., 2007) and the nucleosome remodeling factor BPTF (Wysocka et al., 2006), as well as dozens of candidate H3K4me3 readers. A caveat of the screen is that a few well-defined H3K4me3 readers, for instance the inhibitor of growth (ING) proteins 1–5 (Shi et al., 2006) and recombination activating gene 2 (RAG2) (Matthews et al., 2007), are not identified, probably because they are of low abundance or, in the case of RAG2, not expressed in HeLa cells. Among the candidate hits are the eight members of the histone acetyltransferase (HAT) SAGA complex. Inspection of the SAGA components reveals a potential methyllsyine-binding double tudor domain present within Sgf29, a component of both the SAGA and ATAC HAT complexes. A detailed biophysical analysis demonstrates high affinity and specific binding of the Sgf29 tudor domain to H3K4me3, and Sgf29 is shown to be required for SAGA recognition of H3K4me3.
To study the interacting partners and chromatin occupancy patterns for Sgf29 and the other hits, the authors stably integrate green fluorescent protein (GFP) in frame at the endogenous gene in HeLa cells using bacterial artificial chromosome (BAC) transgenomics. The authors use this system to identify interacting partners of GFP fusions and for deep sequencing of anti-GFP chromatin immunoprecipitates (ChIP-seq). Several important observations are revealed in these analyses. First, all of the H3K4me3 candidate interactors that lack a canonical methyllysine-binding domain interact with proteins (and are potentially members of complexes) that do possess a known H3K4me3 reader, explaining why these hits were specifically pulled down by the H3K4me3 peptides. For example, GATAD1 is identified as an H3K4me3 interactor, and it binds to the lysine demethylase Jarid1A/RBBP2, which contains a PHD finger that recognizes H3K4me3 (Wang et al., 2009). Second, ChIP-seq of GFP fusions to five different H3K4me3 interactors (Sgf29, TRRAP, PHF8, GATAD1, and BAP18) reveals promoter enrichment that correlates with the presence of H3K4me3. The authors also observe considerable overlap in target genes for the five GFP-H3K4me3 inter-actors, demonstrating that multiple different readers can bind a chromatin mark located at a specific genomic site (Figure 1).
Interestingly, Sgf29, TRRAP, PHF8, and BAP18 are likely to be involved in transcriptional activation, whereas GATAD1 probably has a role in transcriptional repression, suggesting that different readers can link H3K4me3 to alternate, even opposing, functions. These results are consistent with previous studies demonstrating that H3K4me3 recognition is essential for readers with diverse functions, including transcription activation, transcription repression, mRNA processing, and V(D)J recombination (Matthews et al., 2007; Shi et al., 2006; Sims et al., 2007; Taverna et al., 2007; Vermeulen et al., 2007; Wang et al., 2009; Wysocka et al., 2006). Thus, the authors provide further evidence that the biological outcome of a specific histone methylation event is a consequence of the protein that recognizes it, rather than being dictated by the modification alone (Figure 1).
The authors observe far fewer proteins interacting with the H3K36me3 peptide relative to H3K4me3 peptide, and no obvious complexes. Notably, four of the most statistically significant H3K36me3 interactors (N-PAC, NSD1, NSD2, and MSH-6) contain a potential methyllsyine-binding PWWP domain, a motif recently reported to bind H3K36me3, albeit with a weak binding affinity relative to other readers (Vezzoli et al., 2010). Vermeulen and colleagues provide evidence that the PWWP domain of GFP-N-PAC is necessary for H3K36me3 recognition. Moreover, they observe enrichment of GFP-N-PAC within the transcribed body of genes, similar to the distribution of H3K36me3. The enrichment of the PWWP domain in the H3K36me3 interactome and the genome-wide distribution pattern of N-PAC argue that PWWP domains may generally recognize H3K36me3, though direct biophysical data is required to definitively categorize the PWWP domain as a bona fide H3K36me3 reader domain. In addition, the low number of hits identified in the H3K36me3 peptide-binding screen may reflect the preference of PWWP domains or other unknown H3K36me3 readers to bind H3K36me3 in the context of nucleosomes rather than peptides. Regardless, it will be insightful to determine whether N-PAC and N-PAC-H3K36me3 interactions play a role in transcription elongation or exon definition, two functions associated with the H3K36me3 mark.
The interactomes identified for the three repressive trimethyllysine histone marks are substantial and include many of the expected known readers. In addition, an intriguing finding is the detection of the origin of replication complex (ORC) in all three peptide pull-downs. The direct molecular link between the ORC complex and binding to H3K9me3, H3K27me3, and H4K20me3 is not elucidated. However, the identification of a new ORC component named LRWD1 may provide a clue. The Poly-comb group protein EED has recently been shown to aid establishment of silent chromatin regions via recognition of repressive trimethyllysine histone marks by its WD40 repeat region (Margueron et al., 2009). A WD40 repeat region is present in LRWD1, suggesting a model analogous to EED in which LRWD1 recognition of H3K9me3, H3K27me3, and H4K20me3 stabilizes ORC at repressive chromatin domains and may therefore promote DNA replication through heterochromatin (Figure 1C).
The recognition of histone methylation by readers has been linked to multiple different functions, including clear evidence that disrupting the readout of a histone modification can cause disease. It is also nearly certain that a vast number of methyllysine-readers with important biological functions await identification, including those that interact with methylated nonhistone proteins. Vermeulen et al. provide a blueprint for how to apply state-of-the-art technology to discover such readers and in doing so establish a framework that promises to deliver new insight into the impact of histone methylation on biology.
References
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, et al. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Hyman AA, Stunnenberg HG, Mann M. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. this issue. [DOI] [PubMed] [Google Scholar]
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Göttgens B, Bycroft M. Nat Struct Mol Biol. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]