Summary
Members of the tristetraprolin (TTP) family of CCCH tandem zinc finger proteins bind to AU-rich regions in target mRNAs, leading to their deadenylation and decay. Family members in Saccharomyces cerevisiae influence iron metabolism, whereas the single protein expressed in Schizosaccharomyces pombe, Zfs1, regulates cell–cell interactions. In the human pathogen Candida albicans, deep sequencing of mutants lacking the orthologous protein, Zfs1, revealed significant increases (> 1.5-fold) in 156 transcripts. Of these, 113 (72%) contained at least one predicted TTP family member binding site in their 3′UTR, compared with only 3 of 56 (5%) down-regulated transcripts. The zfs1Δ/Δ mutant was resistant to 3-amino-1,2,4-triazole, perhaps because of increased expression of the potential target transcript encoded by HIS3. Sequences of the proteins encoded by the putative Zfs1 targets were highly conserved among other species within the fungal CTG clade, while the predicted Zfs1 binding sites in these mRNAs often ‘disappeared’ with increasing evolutionary distance from the parental species. C. albicans Zfs1 bound to the ideal mammalian TTP binding site with high affinity, and Zfs1 was associated with target transcripts after co-immunoprecipitation. Thus, the biochemical activities of these proteins in fungi are highly conserved, but Zfs1-like proteins may target different transcripts in each species.
Introduction
Tristetraprolin (TTP) is a member of a family of tandem CCCH zinc finger (TZF) proteins, which can bind to and destabilize specific mRNAs in vertebrates (Brooks and Blackshear, 2013). The TTP TZF domain interacts with AU-rich elements (AREs) present in target mRNA 3′-untranslated regions (UTRs) and promotes deadenylation and destruction of the mRNA, at least in part by recruiting the Not1 deadenylase complex (Sandler et al., 2011; Fabian et al., 2013). In fungi, proteins containing TZF domains have been studied in both Schizosaccharomyces pombe and Saccharomyces cerevisiae. S. pombe expresses a single protein of this type, named Zfs1 (Kanoh et al., 1995). We identified Zfs1 target mRNAs in this species by comparing transcript levels in wild type (WT) and zfs1Δ mutants (Cuthbertson et al., 2008; Wells et al., 2012). Many transcripts with increased expression in zfs1Δ mutants encoded cell surface glycoproteins known to be involved in cell–cell adhesion (Wells et al., 2012). Some of these appeared to be direct targets of Zfs1, as determined by the presence of Zfs1 binding sites in the transcripts, effects on mRNA decay rates and co-immunoprecipitation of the transcripts with tagged Zfs1 (Wells et al., 2012). These data suggested that a major physiological function of Zfs1 in S. pombe is to regulate the levels of cell surface proteins involved in cell–cell interactions.
Unlike S. pombe, S. cerevisiae expresses two TTP family members, Cth1 and Cth2 (Ma and Herschman, 1995; Thompson et al., 1996). In contrast to the adhesion transcript targets in S. pombe (Wells et al., 2012), S. cerevisiae uses these proteins to regulate a set of transcripts involved in iron metabolism (Puig et al., 2005). However, in S. pombe, the loss of ZFS1 appears to have no effect on the expression of iron metabolism genes (Cuthbertson et al., 2008; Wells et al., 2012). Somewhat surprisingly, the sets of transcripts affected by these proteins in the two organisms appear to be very different. To address this paradox, we evaluated mutants deleted in the single analogous gene, ZFS1, expressed in a third fungal species, Candida albicans.
Candida albicans and several related species are members of the fungal ‘CTG clade’, whose members translate the CUG codon as serine rather than leucine (Santos and Tuite, 1995). Candida species within the CTG clade account for approximately 95% of all identifiable Candida infections (Pfaller and Diekema, 2007). C. albicans is a common constituent of human gut and oral microbiomes that is found in approximately 80% of the population. However, it can cause infections that range from superficial mucosal infections, such as vulvovaginal candidiasis and oral thrush, to severe life-threatening and invasive infections, such as disseminated bloodstream infections. Both superficial and invasive Candida infections can occur in immunocompetent individuals, but are most common and severe in immunocompromised individuals. In addition, C. albicans is the fourth most common cause of hospital-acquired infectious diseases in the United States (Miller et al., 2001). C. albicans is known for its ability to develop drug resistance and evade host defenses, primarily through the formation of biofilms. A biofilm is an organized community of cells adhered to a surface that has distinct characteristics from those of free-living (planktonic) cells, such as enhanced drug resistance and the ability to evade the immune system.
Candida albicans expresses a single, considerably shorter TTP family member, termed Zfs1 (204 amino acids in C. albicans vs. 404 in S. pombe). While CTH1 and CTH2 are up-regulated during iron deprivation, C. albicans ZFS1 is not up-regulated, suggesting that it does not play a role in iron homeostasis (Lan et al., 2004; Puig et al., 2005). However, ZFS1 expression is regulated in C. albicans during the yeast to hyphae switch, a step important for biofilm formation (Nantel et al., 2002). In addition, transcription profiling during biofilm formation has shown that ZFS1 expression increases at specific times during biofilm development, suggesting a potential role in biofilm growth and maturation (Bonhomme et al., 2011). While expression of ZFS1 changes during biofilm development, previous studies have shown that the deletion of ZFS1 had no effect on infectivity, morphogenesis or proliferation in mouse and cell culture models (Noble et al., 2010).
To explore the physiological role of Zfs1 in C. albicans, we evaluated two independent deletion mutant strains for ZFS1. Phenotypic analysis confirmed that zfs1Δ/Δ mutants were viable, with minor alterations in biofilm architecture and slight increases in cell growth during stationary phase. They exhibited no apparent defects in cell–cell interactions. Using mRNA-Seq analysis, we identified 156 transcripts that were significantly increased by 1.5-fold or more in zfs1Δ/Δ mutant strains compared with WT. Of these transcripts, 72% contained the central core of the optimal TTP family member binding site, UAUUUAU. In support of this predicted binding site sequence, recombinant, full-length C. albicans Zfs1 protein bound to the optimal mammalian TTP target sequence, UUAUUUAUU, with nearly identical affinity to the human TTP TZF domain peptide. In addition, using RNA immunoprecipitation (RIP), we found specific interaction of a tagged version of Zfs1 with proposed target transcripts containing the optimum TTP binding sequence. In contrast, of the 56 transcripts that were down-regulated to the same extent, only 5% contained a potential binding site of this type.
The sequences of the proteins encoded by the putative Zfs1 targets were highly conserved among other members of the CTG clade. However, the AU-rich predicted binding sites in these target mRNAs rapidly ‘disappeared’ with increasing evolutionary distance from the parental species. In addition, the majority of proteins encoded by the C. albicans target transcripts did not appear to overlap with those of S. pombe or S. cerevisiae. We propose that the biochemical mechanisms of fungal TTP family member-promoted mRNA decay are similar in these diverse species, but that each organism has evolved its own set of target transcripts to meet its particular physiological requirements.
Results
Phenotype of the zfs1Δ/Δ mutants in C. albicans
Growth
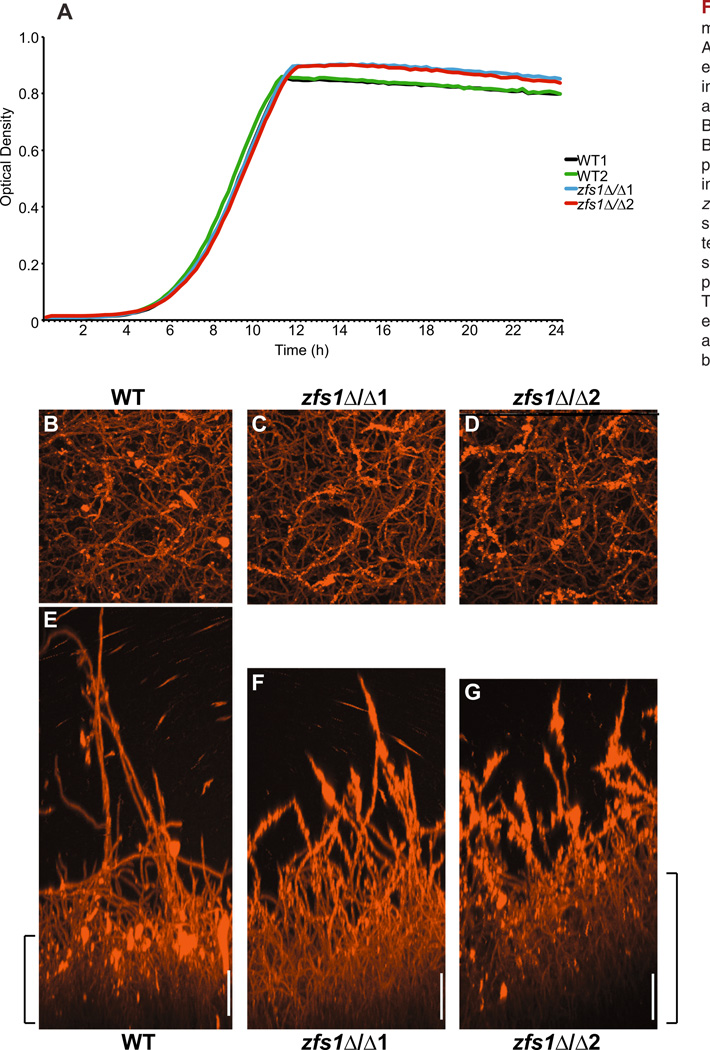
Previous studies in S. pombe have shown that a slight growth defect in zfs1Δ mutants is due to the misregulation of the Wee1 mitotic onset pathway (Navarro and Nurse, 2012). We therefore examined the growth rates of the two independent isolates of C. albicans zfs1Δ/Δ mutants and their corresponding isogenic WT counterparts. Although there were no differences in exponential growth, the loss of ZFS1 resulted in a slight but significant increase in cell density during stationary phase (Fig. 1A). Stationary phase cells are known to have increased adherence, virulence and drug resistance (McCourtie and Douglas, 1984; Mayer et al., 2013), raising the possibility that the loss of ZFS1 may result in an increase in pathogenicity. However, previous studies have demonstrated no effect of Zfs1 deficiency on the virulence of this organism in mouse assays of infectivity, morphogenesis or proliferation (Noble et al., 2010).
Fig. 1. Phenotypic analysis of zfs1Δ/Δ mutants.
A. Growth was measured by optical density every 15 min over a 24 h time period in two independent isolates of wild type (WT1, WT2) and zfs1Δ/Δ mutants (zfs1Δ/Δ1 and zfs1Δ/Δ2).
B–G. Biofilm formation of zfs1Δ/Δ mutants. Biofilm growth was performed on polystyrene plates using wild type (WT) and two independent isolates of zfs1Δ/Δ (zfs1Δ/Δ1, zfs1Δ/Δ2) mutants, and visualized by confocal scanning laser microscopy as described in the text. Upper panels (B–D) for each image show the confocal top view and the lower panels (E–G) show the confocal side view. The substrate is located at the bottom of each image (E–G) and brackets indicate approximate location of the basal layer. Scale bars represent 50 µm.
Biofilm formation
ZFS1 is transcriptionally regulated during the yeast to hyphae switch and is induced during biofilm formation (Nantel et al., 2002; Bonhomme et al., 2011). Therefore, we examined biofilm formation on polystyrene plates in WT and zfs1Δ/Δ mutants. We found a slight, but reproducible, enhancement of the basal biofilm layer on polystyrene plates, consistent with the enhanced cell density we observed during stationary phase (Fig. 1B – G). Because C. albicans biofilm basal layers typically consist predominantly of yeast-form cells, we hypothesized that the zfs1Δ/Δ mutant strain may be defective in hyphal formation. However, we failed to detect a difference in hyphal formation in zfs1Δ/Δ mutants on liquid or solid medium (data not shown).
Flocculation assay
To determine whether the loss of ZFS1 in C. albicans resulted in cell–cell adhesion defects, we analyzed sedimentation rates in the zfs1Δ/Δ mutants. We found that upon the addition of calcium, the zfs1Δ/Δ mutants remained completely in suspension, suggesting that there was no defect in cell–cell adhesion, unlike previous observations in S. pombe (Wells et al., 2012).
Transcript changes in the zfs1Δ/Δ mutants
The minimal growth and viability phenotype exhibited by the zfs1Δ/Δ mutants meant that we could evaluate gene expression changes in the absence of gross morphological abnormalities. Previous studies of Zfs1 in S. pombe and of Cth1/Cth2 in S. cerevisiae have identified several target transcripts that are up-regulated in the deletion mutants. Some of these transcripts were shown to be direct targets of these proteins. However, these mRNA targets were involved in completely different processes within the two species. Therefore, in order to examine the role of Zfs1 in an additional species, we examined changes in gene expression in the zfs1Δ/Δ mutant strains in C. albicans during exponential growth.
C. albicans is a diploid organism that requires two rounds of gene disruption to create a homozygous deletion mutant (Homann et al., 2009).To avoid concerns regarding secondary mutations, we utilized two independent deletion strains for our analyses (Homann et al., 2009). Using mRNA-Seq analysis, we identified 56 transcripts that were significantly (P < 0.05) up-regulated and 10 transcripts (including the deleted transcript) that were significantly down-regulated by at least twofold in both of the zfs1Δ/Δ mutant strains as compared with its isogenic WT strain (Tables 1 and 2). In addition, another 100 transcripts were increased between 1.5- and 2-fold (P < 0.05), and 46 additional transcripts were down-regulated between 1.5 and 2-fold (P < 0.05) (data not shown; data deposited in NCBI Gene Expression Omnibus, Accession Number: GSE53073). We then searched the annotated 3′UTR sequences, as defined by Bruno et al. (2010), of the top 56 transcripts in Table 1 for potential TTP family member binding sites, for which the optimum core sequence in mammals is UAUUUAU. Of the 56 up-regulated transcripts, 44 (79%) contained at least a single 7-mer binding site with this sequence, with many containing overlapping 7-mers and full 9-mers (UUAUUUAUU) (Table 1). Several of the other up-regulated transcripts contained sequences that could represent active binding sites using a less stringent consensus, as illustrated previously for the mammalian TTP TZF domain peptide (Table 1) (Brewer et al., 2006). In contrast, none of the down-regulated transcripts contained a 7-mer potential binding site (Table 2), with the exception of the deleted gene itself. When the 1.5-fold, P < 0.05 cutoffs were used, a total of 156 transcripts were up-regulated in the zfs1Δ/Δ cells, with 113 (72%) containing at least one optimal potential TTP family member binding site. In contrast, using the same cutoffs for down-regulated transcripts, only 3 of the 56 down-regulated transcripts (not including the deleted transcript) contained potential binding sites of this type.
Table 1.
Up-regulated transcripts in zfs1Δ/Δ mutants.
| orf name | Gene | Fold change |
P value | 9-mers | 7-mers | Average WT RPKM |
Function |
|---|---|---|---|---|---|---|---|
| orf19.183 | HIS3 | 15.62 | 1.4E−17 | 7* | 7 | 4.8 | Imidazoleglycerol-phosphate dehydratase, enzyme of histidine biosynthesis |
| orf19.3352 | 12.54 | 3.6E−15 | 6* | 7 | 7.4 | Has domain(s) with predicted oxidoreductase activity | |
| orf19.4028 | 5.33 | 1.3E−07 | 1 | 1 | 36.7 | Putative cis-prenyltransferase involved in dolichol synthesis | |
| orf19.5517 | 4.46 | 3.1E−08 | 0 | 2 | 12.0 | Similar to alcohol dehydrogenases | |
| orf19.558 | GUT1 | 4.35 | 3.3E−07 | 0 | 0 | 15.2 | Putative glycerol kinase |
| orf19.4393 | CIT1 | 4.24 | 8.7E−07 | 3 | 4 | 111.8 | Citrate synthase |
| orf19.3516 | 3.95 | 3.4E−06 | 1 | 2 | 15.0 | Protein of unknown function | |
| orf19.4096 | TAZ1 | 3.93 | 1.3E−05 | 1 | 2 | 3.9 | Putative lyso-phosphatidylcholine acyltransferase |
| orf19.2496 | ATO2 | 3.86 | 6.7E−10 | 3* | 3 | 102.9 | Putative fungal-specific transmembrane protein |
| orf19.5565 | 3.85 | 1.7E−05 | 0 | 1 | 19.2 | Putative 3-hydroxyisobutyrate dehydrogenase | |
| orf19.4600 | 3.65 | 1.7E−08 | 2*/1 | 3 | 8.5 | Protein of unknown function | |
| orf19.136 | 3.59 | 6.1E−08 | 2*/1 | 3 | 5.5 | Predicted membrane transporter | |
| orf19.1224 | FRP3 | 3.24 | 3.2E−07 | 4*/2* | 6 | 93.1 | Putative ammonium transporter |
| orf19.4122 | 3.24 | 4.1E−08 | 8* | 2*/9 | 25.2 | Ortholog(s) have acyl-CoA hydrolase activity | |
| orf19.499 | 3.21 | 4.9E−05 | 3* | 3 | 3.9 | Ortholog(s) have S-adenosylmethionine-dependent methyltransferase activity |
|
| orf19.3391 | ADK1 | 3.1 | 6.1E−05 | 1 | 1 | 333.4 | Putative adenylate kinase |
| orf19.588 | 3.08 | 2.7E−04 | 0 | 1 | 5.2 | Ortholog(s) have role in aerobic respiration and mitochondrial intermembrane space |
|
| orf19.5255 | PXA2 | 3.04 | 1.5E−07 | 0 | 0 | 14.6 | Putative peroxisomal subfamily ABC transporter |
| orf19.1756 | GPD1 | 3.02 | 1.3E−05 | 1 | 2 | 73.1 | Glycerol-3-phosphate dehydrogenase |
| orf19.7411 | OAC1 | 2.9 | 1.1E−04 | 1 | 2 | 2.1 | Putative mitochondrial inner membrane transporter |
| orf19.1340 | 2.82 | 2.4E−05 | 1 | 1 | 28.7 | Putative aldose reductase | |
| orf19.3226 | 2.73 | 1.0E−03 | 0 | 0 | 38.5 | Ortholog(s) have role in intracellular sterol transport and fungal-type vacuole lumen localization |
|
| orf19.1397 | 2.72 | 7.3E−06 | 3/2* | 5 | 7.9 | Has domain(s) with predicted heme binding activity | |
| orf19.6435 | 2.69 | 1.2E−03 | 0 | 1 | 384.7 | Highly conserved subunit of mitochondrial pyruvate carrier | |
| orf19.1618.1 | 2.64 | 1.4E−03 | 0 | 1 | 97.1 | Ortholog(s) have cytosol localization | |
| orf19.1480 | 2.59 | 1.4E−03 | 0 | 3 | 104.3 | Putative succinate dehydrogenase | |
| orf19.6548 | ISU1 | 2.59 | 8.0E−06 | 0 | 1 | 51.7 | Protein with similarity to NifU |
| orf19.5559 | RAV2 | 2.56 | 5.3E−07 | 0 | 0 | 18.9 | Protein similar to Saccharomyces cerevisiae Rav2, a regulator of (H+)-ATPase in vacuolar membrane |
| orf19.5216 | 2.55 | 2.0E−04 | 2*/1 | 3 | 4.6 | Has domain(s) with predicted acyl-CoA hydrolase activity | |
| orf19.2829 | 2.51 | 4.2E−05 | 0 | 0 | 86.2 | Ortholog(s) have role in protein transport and endoplasmic reticulum |
|
| orf19.431 | ZCF2 | 2.44 | 7.1E−06 | 3* | 3 | 32.1 | Zn(II)2Cys6 transcription factor |
| orf19.637 | SDH2 | 2.44 | 7.6E−10 | 1 | 1 | 163.4 | Succinate dehydrogenase |
| orf19.2198 | FLC3 | 2.37 | 7.7E−07 | 3* | 3 | 19.3 | Protein involved in heme uptake |
| orf19.2871 | SDH12 | 2.35 | 4.8E−06 | 0 | 1 | 206.6 | Succinate dehydrogenase |
| orf19.3627 | 2.34 | 1.1E−04 | 1 | 2 | 8.5 | Protein of unknown function | |
| orf19.4895 | 2.33 | 1.7E−04 | 2* | 2 | 3.9 | Protein of unknown function | |
| orf19.6916 | 2.33 | 5.4E−04 | 0 | 0 | 31.9 | Protein of unknown function | |
| orf19.3664 | HSP31 | 2.28 | 5.7E−03 | 0 | 0 | 64.6 | Putative 30 kDa heat shock protein |
| orf19.5921 | 2.25 | 3.7E−04 | 0 | 1 | 11.7 | Protein of unknown function | |
| orf19.1631 | ERG6 | 2.22 | 3.6E−05 | 0 | 0 | 198.3 | Delta(24)-sterol C-methyltransferase |
| orf19.3963 | 2.22 | 1.1E−05 | 0 | 2 | 41.4 | Ortholog(s) have mitochondrion localization | |
| orf19.5711 | 2.19 | 9.7E−09 | 0 | 1 | 26.9 | Putative phosphatidylinositol transfer protein | |
| orf19.3710 | YHB5 | 2.13 | 2.0E−03 | 0 | 1 | 13.6 | Flavohemoglobin-related protein |
| orf19.4575 | 2.12 | 1.9E−06 | 0 | 0 | 15.6 | Ortholog(s) have mitochondrion localization | |
| orf19.5050 | MTO1 | 2.12 | 2.5E−07 | 4* | 4 | 7.3 | Putative mitochondrial protein |
| orf19.4612 | 2.11 | 4.7E−06 | 0 | 0 | 6.0 | Protein with a dienelactone hydrolase domain | |
| orf19.4933 | FAD3 | 2.11 | 1.6E−03 | 3* | 3 | 9.7 | Omega-3 fatty acid desaturase |
| orf19.6938 | MEU1 | 2.11 | 5.8E−05 | 0 | 1 | 26.7 | Putative methylthioadenosine phosphorylase |
| orf19.768 | SYG1 | 2.1 | 6.3E−07 | 1 | 2 | 16.8 | Protein of unknown function |
| orf19.234 | PHA2 | 2.07 | 1.2E−04 | 1 | 1 | 17.5 | Putative prephenate dehydratase |
| orf19.517 | HAP31 | 2.07 | 1.7E−04 | 1 | 1 | 92.3 | CCAAT-binding transcription factor |
| orf19.6724 | FUM12 | 2.05 | 1.2E−04 | 0 | 0 | 69.4 | Putative fumarate hydratase |
| orf19.6805 | 2.05 | 2.9E−04 | 3* | 4 | 5.3 | Protein of unknown function | |
| orf19.685 | YHM1 | 2.05 | 6.5E−04 | 0 | 1 | 79.5 | Putative mitochondrial carrier protein |
| orf19.5589 | 2.01 | 1.6E−02 | 0 | 0 | 24.3 | Protein of unknown function | |
| orf19.6165 | KGD1 | 2.01 | 1.0E−08 | 1 | 2 | 101.9 | Putative 2-oxoglutarate dehydrogenase |
Shown are up-regulated transcripts with average fold changes, P values, average RPKM for wild type (WT) and proposed function for two independent isolates of zfs1Δ/Δ mutants versus WT as determined by mRNA-Seq. The number of 9-mers and 7-mers within the predicted 3′ UTR is also shown. The asterisk (*) indicates overlapping 9-mers or 7-mers.
Table 2.
Down-regulated transcripts in zfs1 Δ/Δ mutants.
| Orf name | Gene | Change | P value | 9-mers | 7-mers | RPKM | Function |
|---|---|---|---|---|---|---|---|
| orf19.5334 | ZFS1 | −12 324.8 | 6.4E−185 | 0 | 4* | 975.1 | Ortholog of Saccharomyces cerevisiae Cth2, an mRNA-binding protein |
| orf19.7585 | INO1 | −2.3 | 5.1E−06 | 0 | 0 | 8.2 | Inositol-1-phosphate synthase |
| orf19.1344 | −2.3 | 4.4E−04 | 0 | 0 | 90.8 | Protein of unknown function | |
| orf19.6993 | GAP2 | −2.3 | 3.4E−05 | 0 | 0 | 8.0 | General amino acid permease |
| orf19.6073 | HMX1 | −2.2 | 1.8E−06 | 0 | 0 | 61.1 | Heme oxygenase |
| orf19.1415 | FRE10 | −2.1 | 8.7E−11 | 0 | 0 | 116.1 | Major cell-surface ferric reductase under low-iron conditions |
| orf19.2602 | OPT1 | −2.1 | 5.0E−08 | 0 | 0 | 88.6 | Oligopeptide transporter |
| orf19.4765 | PGA6 | −2.1 | 1.5E−06 | 0 | 0 | 142.8 | GPI-anchored cell wall adhesin-like protein |
| orf19.6570 | NUP | −2.1 | 1.6E−03 | 0 | 0 | 9.4 | Nucleoside permease |
| orf19.7276.1 | TLO4 | −2.0 | 9.9E−03 | 0 | 0 | 30.4 | Member of a family of telomere-proximal genes |
| orf19.4716 | GDH3 | −2.0 | 9.3E−05 | 0 | 0 | 297.0 | NADP-glutamate dehydrogenase |
Shown are down-regulated transcripts with average fold changes, P values, average RPKM (reads per kilobase of transcript per million reads mapped) for wild type (WT) and proposed function for two independent isolates of zfs1ΔΔ mutants versus WT as determined by mRNA-Seq. The number of 9-mers and 7-mers within the predicted 3′ UTR is also shown. The asterisk (*) indicates overlapping 9-mers or 7-mers.
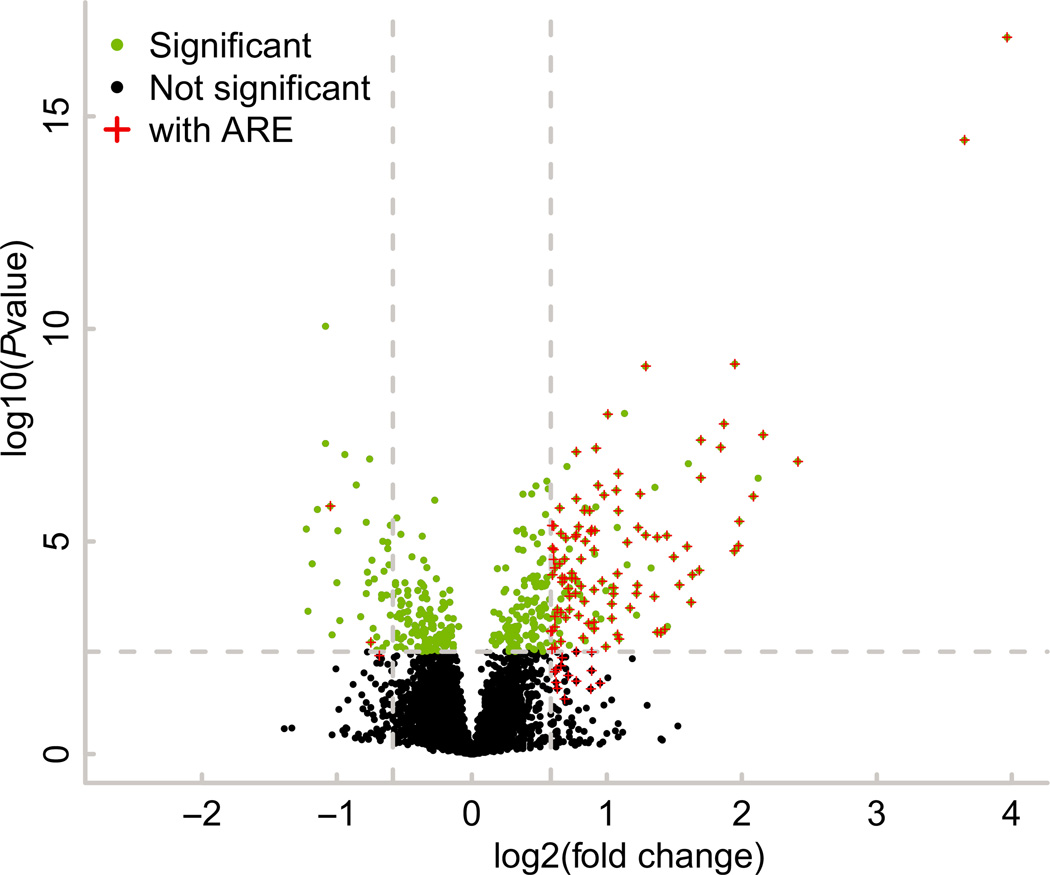
These results are illustrated graphically in Fig. 2, which demonstrates clearly the association between the up-regulated transcripts and the presence of at least one optimal 7-mer binding site. When the percentages of transcripts containing at least one 7-mer potential binding site were compared between the up- and down-regulated transcripts, using Fisher’s exact test, the differences were highly significant using both the 1.5-fold and 2-fold cutoffs (P < 0.0001 in both cases).
Fig. 2.
Up- and down-regulated transcripts that contain at least one potential TTP family member binding site. Shown is a volcano plot for transcripts identified by mRNA-Seq with a raw read count of at least 100. Green dots represent transcripts with a multiple-test adjusted P value ≤ 0.05; this level is also indicated by the horizontal dashed line. Up-and down-regulated transcripts that were changed by at least 1.5-fold are to the right and left of the vertical dashed lines respectively. Transcripts that contain at least a single 7-mer binding site in their 3′UTR are indicated with a red plus (+).
To determine the overall frequency of ARE-containing transcripts in the C. albicans transcriptome, we analyzed the annotated 3′UTR sequences (Bruno et al., 2010). We found that approximately 17% of C. albicans transcripts contained at least a single 7-mer. This is slightly lower than the frequency seen in S. pombe, which contains 22% (Wells et al., 2012). The figure for C. albicans should be viewed as an approximation because of uncertainties in the exact transcript boundaries in this species. However, the data suggest that the high proportion of ARE-containing potential target transcripts identified by mRNA-Seq is not a consequence of an overabundance of ARE-containing transcripts in the C. albicans transcriptome.
To determine whether the Zfs1 targets clustered by specific cellular functions, we performed Gene Ontology (GO) analysis on the top 56 up-regulated transcripts using the Candida Genome Database (CGD) GO Term Analysis tool (http://www.candidagenome.org/cgi-bin/GO/goTermMapper). We found that there was a significant enrichment (6 of 56) of transcripts involved in the tricarboxylic acid (TCA) cycle in our mRNA-Seq dataset, as compared with the transcripts included in the GO dataset (16 of 6525) (P = 4.9E−07) (Table 3). However, we did not find that the down-regulated transcripts were significantly overrepresented in any specific category. Unlike the TTP family members in S. cerevisiae that play roles in iron homeostasis (Puig et al., 2005) or Zfs1 in S. pombe that plays a role in cell–cell adhesion (Wells et al., 2012), we did not observe changes in any transcripts in C. albicans that were shown to be involved in either of these processes, with the exception of FRE10, encoding a major cell-surface ferric reductase under low-iron conditions. However, the ortholog of FRE10 in S. cerevisiae, FRE2, was not found to be a target of Cth1/2 (Puig et al., 2005). In addition, we identified three up-regulated transcripts involved in the TCA cycle, KGD1, SDH2 and CIT1, that had been previously proposed to be Cth1/Cth2 targets (Puig et al., 2005). However, the analysis in S. cerevisiae was performed in iron-depleted conditions while our analysis was performed in iron-rich conditions. To further investigate a possible role in iron regulation, we examined the orthologs of transcripts previously shown to be regulated by Cth1/2 in S. cerevisiae (ACO1, SDH4, HEM15, HEM13 and HMX1) in C. albicans zfs1Δ/Δ mutants using real-time reverse transcription polymerase chain reaction (RT-PCR). We found no changes in expression of these transcripts when we compared WT and zfs1Δ/Δ mutants in either iron-rich or iron-depleted conditions (data not shown). We also found, as shown previously, that neither C. albicans ZFS1 nor S. pombe ZFS1 expression changes significantly during iron deprivation (Cuthbertson et al., 2008; Homann et al., 2009), unlike CTH1/CTH2 in S. cerevisiae, which both increase during iron deprivation.
Table 3.
GO analysis of the mRNA-Seq-identified up-regulated transcripts.
| GO_term | Cluster frequency | Background frequency | P value |
|---|---|---|---|
| Tricarboxylic acid cycle | 6 out of 56 genes | 16 out of 6525 | 4.9E−07 |
| Cellular respiration | 8 out of 56 genes | 95 out of 6525 | 2.6E−04 |
| Oxidation–reduction process | 15 out of 56 genes | 418 out of 6525 | 3.0E−04 |
| Aerobic respiration | 7 out of 56 genes | 71 out of 6525 | 4.4E−04 |
| Energy derivation by oxidation of organic compounds | 8 out of 56 genes | 118 out of 6525 | 1.3E−03 |
| Single-organism metabolic process | 23 out of 56 genes | 1073 out of 6525 | 2.1E−03 |
| Generation of precursor metabolites and energy | 8 out of 56 genes | 135 out of 6525 | 3.6E−03 |
| Monocarboxylic acid transport | 4 out of 56 genes | 25 out of 6525 | 1.2E−02 |
| Small molecule metabolic process | 15 out of 56 genes | 633 out of 6525 | 4.3E−02 |
A CGD GO term analysis is shown of the 56 up-regulated transcripts identified by mRNA-Seq along with the specific function, the cluster frequency, the background frequency and the P value. The cluster frequency is the number of genes identified by mRNA-Seq within our dataset that are assigned to a specific GO function. The background frequency is the number of genes identified in a specific GO function within the Candida albicans transcriptome.
Zfs1 target transcripts in C. albicans
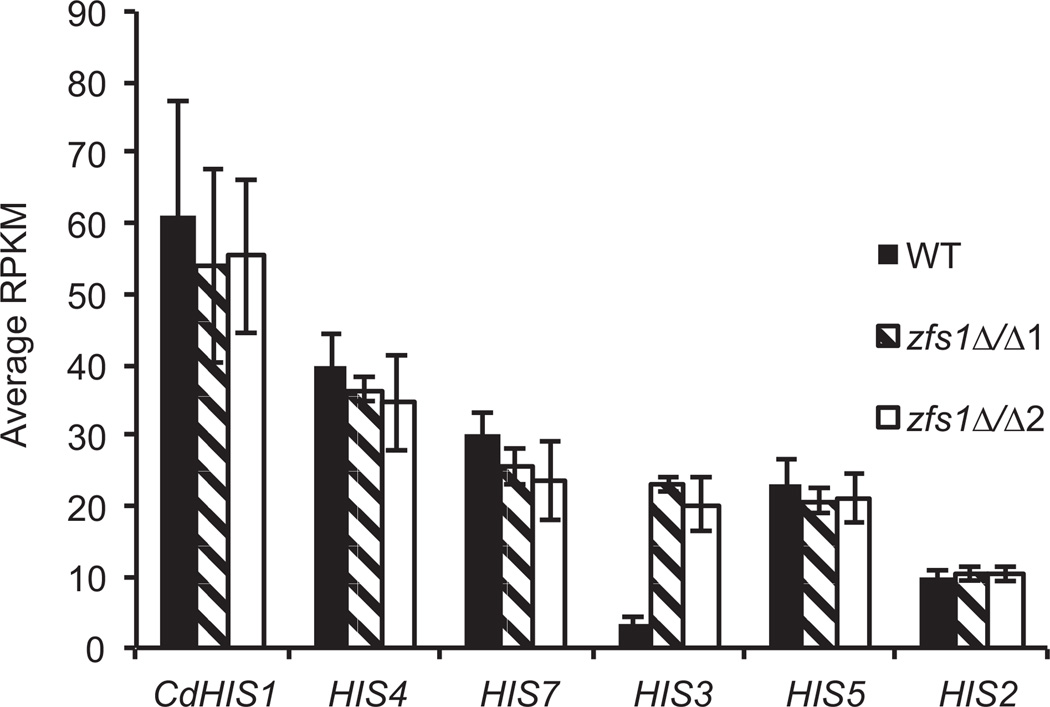
Interestingly, the top potential target identified by mRNA-Seq (15.6-fold up-regulated, P =1.4E−17) is orf19.183 (HIS3), which encodes the imidazoleglycerol-phosphate dehydratase enzyme involved in histidine biosynthesis (Table 1). To verify that the increase in HIS3 expression observed in zfs1Δ/Δ mutants was not due to the use of Candida dubliniensis HIS1 as the deletion cassette, we examined the transcript levels for the enzymes of the histidine biosynthesis pathway (Gomez-Raja et al., 2008). We found no significant changes in any of the histidine pathway transcripts, with the exception of the HIS3 mRNA itself (Fig. 3). In addition, the C. dubliniensis HIS1 was also integrated into the genome of the WT strain used in our analysis. Taken together, the increase in HIS3 mRNA observed in the zfs1Δ/Δ mutants most likely reflects direct regulation of HIS3 mRNA turnover rather than an overall perturbation of the pathway.
Fig. 3.
Expression of genes within the histidine biosynthesis pathway. Shown are the average RPKM values ± standard deviation from wild type, zfs1Δ/Δ1 and zfs1Δ/Δ2 strains, for the indicated transcripts in the histidine biosynthesis pathway. CdHIS1 represents HIS1 from Candida dubliniensis that was used for disruption of one allele of ZFS1 in the zfs1Δ/Δ strains, and was also inserted into the isogenic wild-type strain.
Previous studies have shown that the HIS3 mRNA 3′UTR contains a microsatellite locus consisting of a repetitive element (ATTT)n that is used in the classification of C. albicans subtypes (Botterel et al., 2001). In addition, disease-causing strains obtained from patients have demonstrated widely varying lengths of this repeat element in the 3′UTR, and these length differences have been suggested to play a role in pathogenicity (Botterel et al., 2001; L’Ollivier et al., 2012). Sequence analysis of the two independent zfs1Δ/Δ strains used here and their isogenic WT control strain showed no differences in the numbers of ATTT repeat elements within the HIS3 3′UTR. Specifically, the length of the HIS3 3′UTR in the GenBank reference strain SC5314 (AACQ01000253) was 67 bases containing 8 overlapping 7-mers, while the HIS3 3′UTR in our strains was 85 bases containing 19 overlapping 7-mers.
Some of the potential target transcripts found in the three fungi studied to date are unique to each organism, while some are fungal-specific and others have apparent orthologs in non-fungal eukaryotes. It was not always possible to find direct orthologs of all C. albicans up-regulated transcripts in S. pombe or S. cerevisiae by protein searching. However, we did not find any overlaps, with the possible exceptions of the three transcripts involved in the TCA cycle, between the lists of putative targets in the three organisms.
Characterization of Zfs1 targets
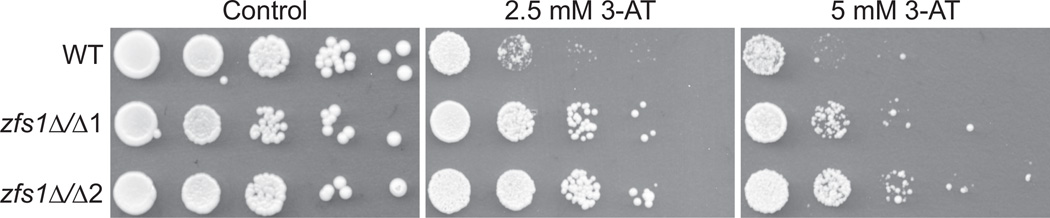
Because of the lack of an obvious phenotype of the zfs1Δ/Δ mutants, we chose three of the most highly up-regulated transcripts that contained potential TTP family member target sequences and examined conditions in which they might be expected to play a functional role. In one case, we tested the hypothesis that increased levels of HIS3 mRNA might allow for growth in 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of imidazoleglycerol-phosphate dehydratase, the product of HIS3 (Bai and Elledge, 1997). We found that the zfs1Δ/Δ mutants were resistant to increasing doses of 3-AT when grown in minimal medium (Fig. 4), suggesting that increased expression of HIS3 mRNA and presumably protein in the mutant could overcome the growth inhibitory effect of 3-AT under these conditions.
Fig. 4.
Growth of zfs1Δ/Δ mutants on 3-AT. Wild type, zfs1Δ/Δ1 and zfs1Δ/Δ2 strains were spotted in 10-fold serial dilutions onto minimal medium plates lacking histidine and containing 0 mM (control), 2.5 mM or 5 mM 3-amino-1,2,4-triazole (3-AT).
A second highly up-regulated transcript, from orf19.3352, is likely to encode an ortholog of S. cerevisiae Env9, a protein involved in vacuolar morphology (Ricarte et al., 2011). To examine vacuolar morphology in zfs1Δ/Δ mutants, we stained cells with FM4-64, a fluorescent styryl dye that selectively stains vacuole membranes in live cells (Ricarte et al., 2011). We observed no differences in vacuole morphology in zfs1Δ/Δ mutants as compared with the WT strain (data not shown). The third test transcript, from orf19.4393 (CIT1), encodes a citrate synthase; increases in this enzyme may affect the growth response to alternative nutrient sources or hypoxia. The GO analysis also showed enrichment of up-regulated transcripts involved in the TCA cycle, suggesting possible increased growth on alternative carbon sources. However, we found no differences in the growth of zfs1Δ/Δ mutants on lactate, galactose, glycerol, sucrose or sorbitol, compared with the WT strain. We also found no differences in growth between WT and zfs1Δ/Δ strains under hypoxic conditions (data not shown). These data suggest that the increase in CIT1 expression does not result in an obvious growth advantage on alternative carbon sources, at least under our conditions.
Finally, we subjected the zfs1Δ/Δ mutants to additional cellular stresses and found no differences in the growth of zfs1Δ/Δ mutants as compared with WT on chemicals that stress the cell wall, such as Congo red or calcofluor white, or the cell membrane, such as dimethyl sulfoxide or for-mamide. In addition, there were no differences in the growth of the zfs1Δ/Δ mutants under conditions that cause oxidative stress, such as hydrogen peroxide (data not shown).
Zfs1 target conservation
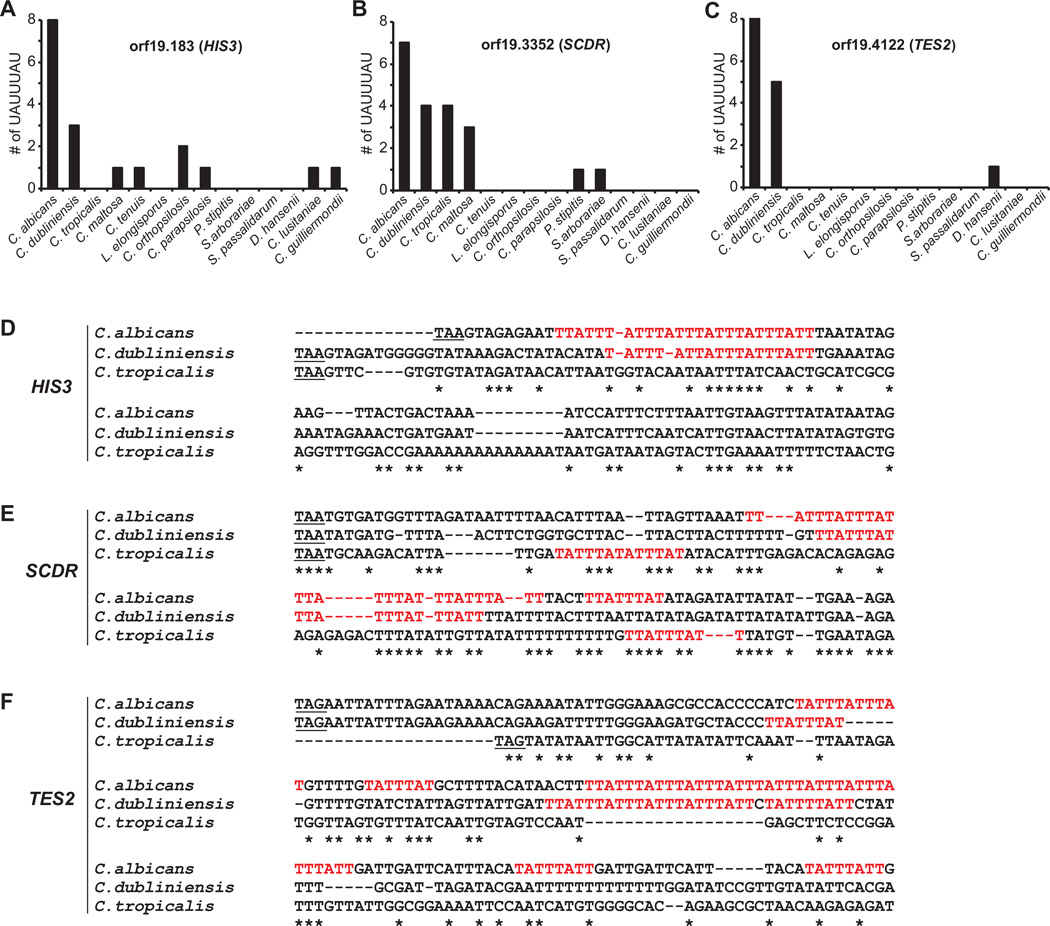
Zfs1 protein-coding sequences are present in at least 13 members of the fungal CTG clade, in addition to C. albicans, that have reasonably complete genomic sequences in GenBank. The Zfs1 alignment shows that the proteins are highly conserved within the C-terminal tandem zinc finger domain (see below), but lose sequence conservation outside of that domain. To investigate whether the ARE binding sites in our proposed target sequences from C. albicans were conserved in other members of the CTG clade, we identified protein orthologs in other CTG clade species of the top three target mRNAs identified by mRNA-Seq that contained at least six or more 7-mer binding sites, orf19.183, orf19.3352 and orf19.4122. Using the stop codons of the predicted proteins as a guide to the 3′UTR, we searched 500 base pairs following the stop codon from each species for 7-mer binding sites. We found 6–12 7-mer binding sites within the HIS3 3′UTR sequences from the 4 different genomic sequences representing 4 different C. albicans sequences currently available in GenBank (AVAX01000022, AACQ01000253, AVAZ01000065, AVAW01000080). When we compared the number of 7-mer binding sites from C. albicans with orthologs in other available members of the CTG clade, we found that the numbers of 7-mer binding sites generally decreased with increasing evolutionary distance from C. albicans (Fig. 5A – C). In fact, the orthologs in several species sometimes had no binding sites whatsoever (Fig. 5A – C).
Fig. 5.
Evolutionary relationships of Zfs1 targets within members of the CTG clade. Shown are the numbers of potential 7-mer binding sites within 500 base pairs of the stop codons in (A) orf19.183, (B) orf19.3352 and (C) orf19.4122. The order of species along the X-axis roughly reflects increasing evolutionary distance of the species from Candida albicans, based on previous phylogenetic comparisons. (D–F) Alignments performed with Clustal Omega of the sequences of the 3′UTRs of (D) HIS3, (E) SCDR and (F) TES2 from the indicated CTG clade species are shown, including the potential binding 7-mers in red. The alignment begins at the stop codon (underlined) and continues at least 60 bases after the 3′-most 7-mer identified in any of the species. Asterisks (*) indicate sequence identity.
A striking demonstration of this phenomenon can be seen in a comparison of the 3′UTR sequences of the same three transcripts in the closely related species C. albicans, C. dubliniensis and Candida tropicalis (Fig. 5D – F). In evolutionary terms, both C. albicans and C. dubliniensis are thought to have C. tropicalis or a closely related species as a common ancestor (McManus and Coleman, 2014). Although the number of 7-mer potential binding sites is decreased in C. dubliniensis compared with C. albicans in all three cases, it is clear that many of these target sequences are at least partially conserved. However, in C. tropicalis, a diploid CTG clade member that has been estimated to exhibit 70% amino acid identity with C. albicans in 5254 orthologous proteins (Butler et al., 2009), there were no potential binding sites within two of the three transcripts (Fig. 5E and F).
We applied the same approach to the top 56 up-regulated transcripts listed in Table 1. Of these transcripts, 44 contained at least one 7-mer potential binding site. We identified the encoded orthologous proteins from C. tropicalis and Lodderomyces elongisporus, the latter being a closely related diploid member of the CTG clade that is not in the direct line of descent from C. tropicalis to C. albicans (McManus and Coleman, 2014). We examined 500 bases downstream of the stop codon for each orthologous protein in the genomic sequences currently available in GenBank. Compared with 44/44 (100%) of the transcripts that contained at least one 7-mer in C. albicans, 33/44 (75%) contained 7-mers of this type in C. tropicalis and 12/44 (27%) contained at least one such 7-mer in L. elongisporus. These data support the concept that with increasing evolutionary distance, there is a decreased frequency of binding sites from individual orthologous transcripts. In most cases in which one or more binding sites were present in the L. elongisporus orthologs, there were decreased numbers of 7-mer potential binding sites. However, in one case, C. albicans orf19.1340, encoding a putative aldose reductase, there was an increase from one 7-mer in C. albicans to eight overlapping 7-mers in L. elongisporus.
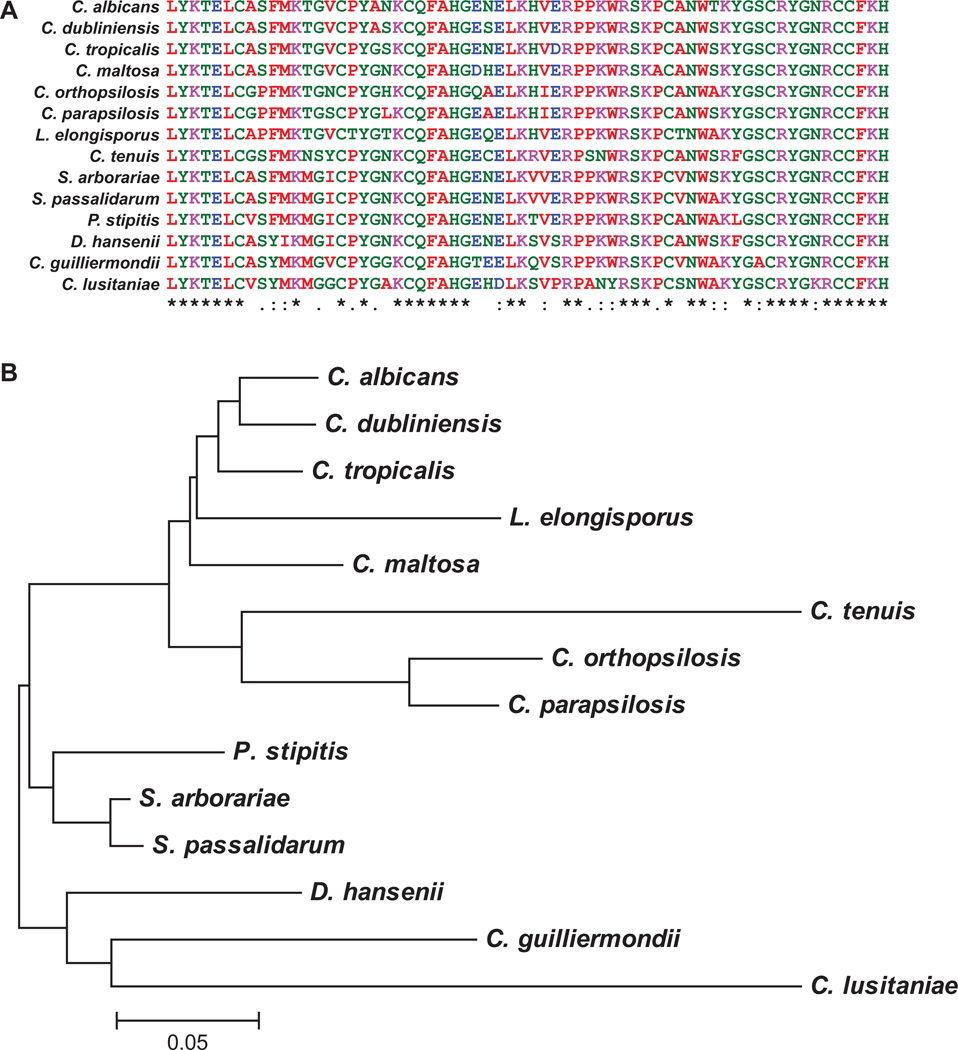
Conservation of the TZF domain in CTG clade species
An alignment of the C. albicans Zfs1 TZF domain sequence with those from the available members of the CTG clade species demonstrates perfect conservation of intra- and inter-finger spacing in all species, as well as conservation of the two pairs of CCCH zinc finger residues and their lead-in sequences (Fig. 6A). We analyzed the relatedness of the TZF domains from these proteins within members of the CTG clade by constructing a phylogenetic tree, using the neighbor-joining method in MEGA5.1 (Fig. 6B) (Tamura et al., 2011). This tree, although constructed using only the 64-amino acid RNA-binding domain, exhibited a very similar structure to ones based on phylogenetic relationships (Butler et al., 2009). The presence or absence of binding sites in Zfs1 targets correlated well with the overall relatedness of the Zfs1 TZF domain sequences (Figs 5 and 6B).
Fig. 6. Conservation of the TZF domain from Zfs1 across CTG clade species.
A. Shown is a sequence alignment of the TZF domain from Candida albicans Zfs1 and from the orthologous protein sequences from the indicated members of the CTG clade. Sequences were aligned by ClustalW2, and colors were assigned by ClustalW2 based on their physiochemical properties. Asterisks (*) indicate amino acid identity at that site; colons (:) indicate a conserved substitution; and dots (.) indicate a semi-conserved substitution at that site.
B. Shown is a phylogenetic tree, demonstrating the relatedness of the Zfs1 TZF domains within species of the CTG clade. This tree used only the TZF domain sequences shown in A. The sequence for each protein was obtained from GenBank as described in the Experimental procedures section, and sequence relationships were determined using the neighbor-joining method (Saitou and Nei, 1987). The original alignment was performed in ClustalW2, and the tree was constructed in MEGA5.1. The evolutionary distances were computed using the Dayhoff matrix based method (Schwartz and Dayhoff, 1979). The tree is drawn to scale, with branch lengths representing the number of amino acid substitutions per site.
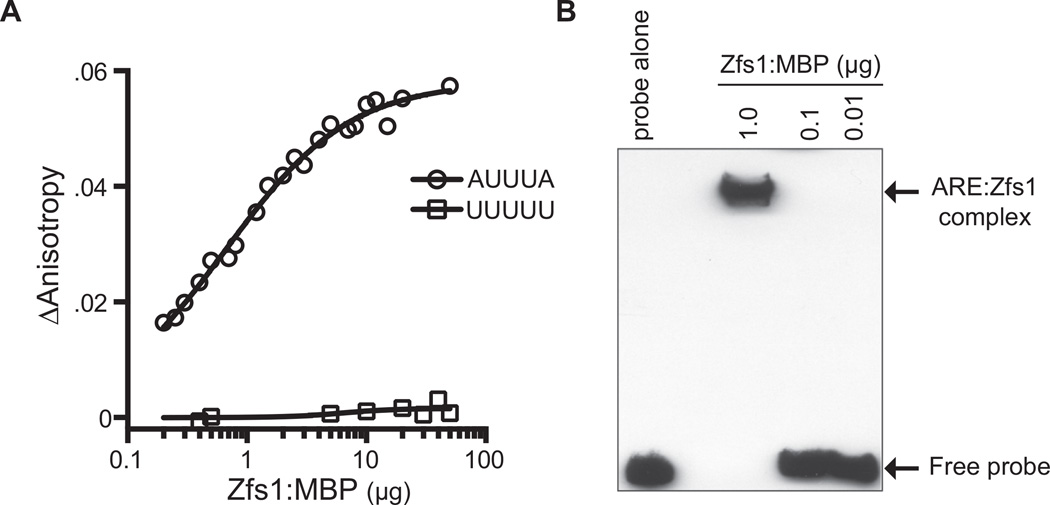
Characteristics of binding of Zfs1 to an ARE RNA oligonucleotide
Previous studies have shown high affinity binding between a 73-amino acid synthetic peptide comprising the TZF domain of human TTP, termed TTP73, and RNA targets containing ARE binding sequences (Brewer et al., 2004). Specifically, the human peptide bound to the optimum TTP binding sequence UUAUUUAUU with a Kd of 3.2 nM at 24°C (Brewer et al., 2004). Deviations from this optimum sequence resulted in decreases in binding affinity (Brewer et al., 2004). To determine whether the C. albicans protein could bind to similar RNA sequences with similar high affinity, we expressed the recombinant protein in Escherichia coli and used the purified protein for binding measurements to a similar RNA oligonucleotide, using fluorescence anisotropy under steady state conditions. These studies used the 13 base fluorescein-labeled RNA substrate (5′-FL-UUUUAUUUAUUUU-3′;FL-ARE13) (Brewer et al., 2004) and the purified C. albicans Zfs1 full-length protein with a Maltose Binding Protein (MBP) tag fused to the N-terminus. Strikingly, similar to the human TTP73 peptide, the purified full-length Zfs1:MBP fusion protein bound with high affinity (Kd of 1.8 nM at 24°C) to the FL-ARE13 probe, with a stoichiometry of 1:1 for the protein-RNA complex (Fig. 7A). We observed essentially no binding of the purified Zfs1:MBP fusion protein to a control polyU probe (5′-FL-UUUUUUUUUUUUU-3′) (Fig. 7A). No fluorescence quenching was observed over the course of the experiments (data not shown). We also performed gel shift analyses, using varying concentrations of purified Zfs1:MBP and a tumor necrosis factor (TNF)-ARE-based RNA probe (Kedar et al., 2012). Incubation of 1.0 µg of the purified Zfs1:MBP fusion protein with the probe resulted in the complete shifting of the probe into a single band complex, whereas no shift was detectable at lower protein concentrations (Fig. 7B).
Fig. 7. Measurement of Zfs1 RNA binding affinity using fluorescence anisotropy.
A. Binding reactions containing the 13 base fluorescein-labeled RNA target (5′-FL-UUUUAUUUAUUUU-3′; FL-ARE13), or the control polyU probe (5′-FL-UUUUUUUUUUUUU-3′), and a titration of purified Zfs1:MBP were mixed and fluorescence intensity was monitored. A nonlinear regression algorithm in PRISM was used to calculate Zfs1-dependent changes in anisotropy. ΔAnistropy was calculated as anisotropy for probe alone subtracted from total anisotropy measured at each protein concentration.
B. Increasing concentrations of purified Zfs1:MBP or no protein (probe alone) were used in a gel shift analysis with a 5′biotin labeled TNF-ARE probe. Arrows to the right indicate migration positions of the ARE probe and the Zfs1:MBP ARE probe complex.
These data demonstrate that the optimum binding site found in studies of human TTP, UUAUUUAUU, is also a high-affinity binding site for the full-length C. albicans protein. Further studies with modified oligonucleotides will be necessary to determine whether the target site sequence specificity is similar between the two organisms.
RIP of Zfs1 targets
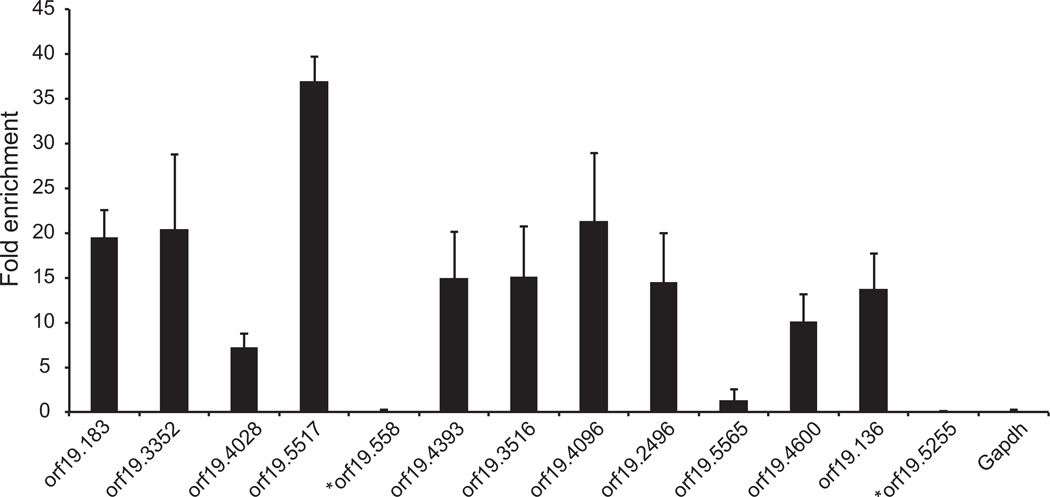
To determine whether Zfs1 could directly interact with the proposed target transcripts, we performed RIP using a strain that expresses a single copy of a fusion protein of Zfs1 with Myc tags that was integrated into the endogenous locus. Following immunoprecipitation of Zfs1:Myc with the Myc antibody, we found that 10 of 11 of the putative target transcripts tested were enriched in the immunoprecipitated RNA from the Zfs1:Myc strain as compared with the control strain lacking the Myc tag (Fig. 8). The single transcript that was not pulled down by the Myc antibody was orf19.5565, which contains only a single 7-mer potential binding site. There was no significant enrichment of either orf19.558 or orf19.5525, both transcripts that were increased in zfs1Δ/Δ mutants but that lacked obvious binding sites (Fig. 8). There was also no enrichment of GAPDH mRNA, used as another negative control (Fig. 8).
Fig. 8.
RNA immunoprecipitation of Zfs1 targets. RNA immunoprecipitations were performed using a strain expressing a C-terminal Myc-tagged Zfs1 and a no tag control strain (SN250). Shown are the mean values of the fold enrichment of Zfs1:Myc immunoprecipitated RNA and ± standard deviation from three independent experiments. The asterisks indicate transcripts that were up-regulated in the Zfs1-deficient cells, but did not contain 7-mer potential TTP family member binding sites.
Discussion
Because the zfs1Δ/Δ mutants lacked an obvious growth or morphological phenotype, we could investigate the biochemical functions of Zfs1 as a potential mRNA-binding and destabilizing protein in the absence of major phenotypic changes. By analogy with the activities of TTP family members in other organisms, we anticipated that direct mRNA targets of Zfs1, the single TTP family member protein expressed in this species, would accumulate in these mutants. Using mRNA-Seq, we identified 56 transcripts that were significantly up-regulated at least twofold in the absence of Zfs1, with another 100 transcripts significantly elevated by 1.5- to 2-fold. Of the 156 transcripts that were up-regulated more than 1.5-fold, 72% contained at least one version of the core mammalian TTP family member binding site sequence, UAUUUAU, whereas only 5% of transcripts down-regulated to the same extent contained similar 7-mer sequences. Using co-immunoprecipitation, we observed specific interactions between Zfs1 and many of the putative target transcripts that contained TTP family member binding sites of this type, but no interaction with up-regulated transcripts lacking the binding site. Finally, we showed that recombinant Zfs1 could bind to a synthetic RNA ‘target’ containing the optimal mammalian binding sequence with low nanomolar affinity, similar to that seen with a human TTP binding domain. These findings support the hypothesis that Zfs1 is acting to destabilize a significant number of transcripts in this organism, and that these accumulate in its absence in a characteristic pattern.
In both S. cerevisiae and S. pombe, the TTP family member proteins seem to act like their mammalian counterparts, in that they can bind to ARE sequences in mRNA and promote the decay of those mRNAs. These studies have relied on the assumption that the proteins’ target specificities are conserved between the mammalian proteins and the fungal proteins, an assumption that has been supported by several types of mutation analysis (Cuthbertson et al., 2008; Vergara et al., 2011). We were able to demonstrate directly that the full-length Zfs1 protein of C. albicans can bind to the optimal mammalian target sequence, UUAUUUAUU, with very nearly identical affinity to that seen with a purified synthetic TZF domain peptide from human TTP, using the same fluorescence anisotropy assay. This is despite many differences in the sequences of the TZF domain from the various CTG clade species compared with human TTP. In future experiments, it will be important to determine whether the affinities of the Candida protein for binding site variants exhibit the same target specificity as the human TZF peptide (Brewer et al., 2004). Although a common ancestor of fungi and mammals is thought to have existed more than a billion years ago (Stajich et al., 2009), it is remarkable that this RNA binding module appears to have been maintained essentially intact during that evolutionary time.
Several of the proposed Zfs1 target transcripts in C. albicans encoded proteins previously identified as playing a role in cell adhesion and pathogenicity. However, unlike the situation in S. pombe, the zfs1Δ/Δ mutants in C. albicans did not appear to have defects in cell–cell adhesion, as evidenced by the lack of flocculation both in normal growth conditions and after calcium treatment. Similarly, in contrast to S. cerevisiae, absence of Zfs1 in S. pombe did not seem to have an effect on iron metabolism (Cuthbertson et al., 2008; Wells et al., 2012) and the potential Zfs1 targets observed in the present study are not obviously part of an iron-related regulon. At first glance, therefore, it appears that the three organisms have used their specific TTP family member proteins for their own specific purposes, which differ from one species to the next.
We explored this concept further by investigating the presence of Zfs1 and its potential target sequences in other members of the CTG clade, whose ancestors are thought to have diverged from other related species approximately 190 million years ago. We found orthologs of Zfs1 in 13 species, and each species appeared to express only a single member of this protein family. We then investigated the conservation of target sequences in the 3′UTRs of the most elevated transcripts in the C. albicans zfs1Δ/Δ mutants. As expected, the proteins encoded by the putative target mRNAs were highly conserved among other CTG clade species, but the predicted 3′UTRs were much less well conserved. Although remnants of the characteristic AU-rich target sequences were generally found in the species closest to C. albicans in evolutionary terms, all traces of these target sequences were often lost in the less related species. In our view, this is a remarkable example of a protein that is apparently highly conserved in presumed biochemical function during evolution, while its specific mRNA targets have evolved and diverged with speciation.
The ‘top’ target identified by our mRNA-Seq analysis was the HIS3 mRNA. The HIS3 gene is known to have polymorphic alleles that encode varying numbers of repeats of the microsatellite DNA sequence (ATTT)n that are located within the 3′UTR of the transcript (Botterel et al., 2001). These repeat numbers have been used to categorize C. albicans clinical isolates into specific subtypes. One study showed that HIS3 microsatellite lengths differed significantly between commensal isolates and clinical disease isolates, and that these markers were stable through several generations (Bart-Delabesse et al., 2001). The precise role, if any, of these sequences in pathogenicity is unknown. Given that the microsatellites provide a series of consecutive and overlapping Zfs1 binding sites, it seems possible that the stability of the HIS3 mRNA differs between transcripts with varying numbers of repeats, and that Zfs1 could thus differentially regulate the expression of different HIS3 alleles. This speculation is supported by previous experiments in mammalian systems, in which increased numbers of binding sites led to increased TTP-dependent mRNA decay (Lai et al., 2005). To date, our attempts to assay the transcripts from two different alleles of widely different microsatellite lengths in the same C. albicans cells have not demonstrated obvious differences in steady state transcript levels, but some form of differential regulation based on microsatellite length remains a possibility.
Our study relied on increases in steady state levels of mRNAs in the zfs1Δ/Δ mutants as an indicator that those mRNAs are likely to be direct targets of Zfs1. These changes occurred in many transcripts that contained potential Zfs1 binding sites, and also occurred in the absence of a gross phenotype in the deletion mutants, which might alter gene expression independently. According to this formulation, the down-regulated transcripts, which were fewer in number and rarely contained potential Zfs1 binding sites, are likely to be secondary effects, perhaps related to one or more of the transcription factors that were up-regulated in the zfs1Δ/Δ cells. Altogether, we found 212 transcripts whose steady state levels were changed by more than 1.5-fold with P < 0.05. Because our mRNA-Seq analysis was able to quantitate results from 5585 transcripts, this indicates that the zfs1Δ/Δ mutation resulted in major expression changes in approximately 4.6% of the C. albicans transcriptome.
Taken together with the data from S. pombe and S. cerevisiae, the data in C. albicans support the concept that the biochemical functions of the TTP family members in these species, i.e. their ability to bind AREs in mRNA with high affinity and promote mRNA decay, are highly conserved in these diverse species. However, while the biochemical functions of the TTP family members may be conserved, target identity is not as well conserved. In fact, even within the members of the CTG clade, as seen here, and previously in members of the Schizosaccharomyces genus (Wells et al., 2012), the TTP family member binding sites within the 3′UTR are often not conserved within the orthologous transcripts. These data suggest that while the basic biochemical function of TTP family members may be conserved throughout evolution, each organism has evolved its own set of transcripts that can be regulated by members of this protein family.
Experimental procedures
Yeast strains and media
All strains are in isogenic backgrounds and all but one were generous gifts from Dr. Alexander Johnson, University of California, San Francisco (UCSF) (Noble et al., 2010). Two independent zfs1 homozygous deletion mutant strains (zfs1Δ/Δ1, zfs1Δ/Δ2) were used in this study. These strains were previously constructed, by homologous recombination using HIS1 from C. dubliniensis and LEU2 from Candida maltosa (Noble et al., 2010). Comparisons were made with the isogenic WT strain, SN250 (Noble et al., 2010). Unless indicated, strains were grown in yeast extract peptone dextrose medium (YPD) at 30°C. The C-terminal tagged Zfs1:Myc strain was constructed in this study following the method described in Nobile et al. (2009):
SN250 – ura3Δ::λimm434::URA3-IRO1/ura3Δ::λimm434; arg4::hisG/arg4::hisG; his1::hisG/his1::hisG; leu2:: hisG::CdHIS1/leu2::hisG::CmLEU2
zfs1Δ/Δ1 – ura3Δ::λimm434::URA3-IRO1/ura3Δ:: λimm434; arg4::hisG/arg4::hisG; his1::hisG/his1::hisG; zfs1::CdHIS1/zfs1::CmLEU2
zfs1Δ/Δ2 – ura3Δ::λimm434::URA3-IRO1/ura3Δ:: λimm434; arg4::hisG/arg4::hisG; his1::hisG/his1::hisG; zfs1::CdHIS1/zfs1::CmLEU2
Zfs1:Myc – ZFS1-Myc-FRT-SAT1-FRT::ZFS1; ura3Δ:: λimm434::URA3-IRO1/ura3Δ::λimm434; arg4::hisG/ arg4::hisG; his1::hisG/his1::hisG; leu2::hisG::CdHIS1/ leu2::hisG::CmLEU2
mRNA-Seq analysis
Total RNA was isolated from four independent cultures of WT and four independent cultures of the two zfs1Δ/Δ mutants, grown on separate days, in YPD during mid-log phase growth, using the RiboPure Yeast RNA Purification kit (Life Technologies) following the manufacturer’s protocol. The RNA was quantitated with a NanoDrop 2000C, and 10 µg of each sample was reverse transcribed using oligo dT primers to generate cDNA libraries, and sequenced at the NIH Intramural Sequencing Center (http://www.nisc.nih.gov) using 36 bp single-end reads on the Illumina Genome Analyzer IIx (Illumina), as described previously (Wells et al., 2012). Both the C. albicans genomic sequence and gene annotation data for the analysis were downloaded from CGD (http://www.candidagenome.org). We used the ELAND alignment tool from Illumina CASAVA software to map mRNA-Seq reads to the whole genome reference (Assembly 21) of C. albicans SC5314. We then used the EpiCenter tool (Huang et al., 2011) to identify differentially expressed genes between different groups. Specifically, in gene read counting, we excluded all reads with a mapping quality score of 0 in order to filter out reads that were poorly aligned or aligned to multiple locations equally well. Read counts were then normalized to the mean of total reads of individual samples in order for read counts of a gene to be compared across samples. Results from one of the zfs1Δ/Δ samples did not pass quality control, so the mean comparisons discussed here used four WT and seven zfs1Δ/Δ samples. When the raw reads greater than 0 from the zfs1Δ/Δ samples were compared with each other, the minimum correlation coefficient between any two samples was 0.96. In addition, when the average reads from the two zfs1Δ/Δ strains were compared, only one relatively poorly expressed transcript (FDH1) was statistically different at 1.68-fold. These findings supported the averaging of read counts from all seven zfs1Δ/Δ samples. All genes with a normalized read count < 100 in both WT and zfs1Δ/Δ groups were then filtered out. Differentially expressed genes were then identified using EpiCenter’s Max-P statistic, which estimates the genome-wide variation of expression levels of all genes from biological replicates within a group, and then uses it to determine the significance of expression change in a gene between two groups.
Volcano plot
The volcano plot was generated by our own program using the R project for statistical computing language (http://www.r-project.org/).
ARE identification
Based on the mRNA-Seq analysis from Bruno et al. (2010), we extracted all annotated 3′UTR sequences. We created a customized program to count the number of non-overlapping 7-mers (UAUUUAU) in each 3′UTR by the simple exact match approach.
Serial dilution assays
For all conditions, strains were grown in YPD to mid-log phase and 10-fold serial dilutions of each strain were spotted onto the following plates and incubated at 30°C for 2 days. For growth on 3-AT, strains were spotted onto minimal medium plates lacking histidine and containing 0 mM (control), 2.5 mM or 5 mM 3-AT (Sigma Aldrich). To test various carbon sources, strains were spotted onto yeast peptone (YP) plates containing 2% lactate, galactose, glycerol, sucrose or sorbitol. To examine cell wall defects, strains were spotted onto YPD plates with a range of 10–30 µg ml−1 of calcofluor white, a range of 50–250 µg ml−1 of Congo red, 7% dimethyl sulfoxide (DMSO) or 4% formamide (all from Sigma Aldrich). For growth under hypoxic conditions, strains were spotted onto YPD plates and placed in an airtight chamber that was continuously filled with 99.9% N2. To examine oxidative stress, strains were spotted onto YPD plates with 6.5 mM hydrogen peroxide.
Vacuole staining
Strains were grown in YPD to mid-log phase and stained with FM4-64 (Life Technologies) as described previously (Ricarte et al., 2011).
Sequence searches and alignments
The protein sequences for orf19.183, orf19.3352 and orf19.4122 were used to search for orthologs in additional species within the CTG clade. The full-length protein sequences were used to search either the reference genome or the whole-genome shotgun contigs database in GenBank using tblastn. We then searched 500–1000 base pairs after the stop codon for each open reading frame. In one case, we used the same strategy to search the 3′UTRs of C. tropicalis and L. elongisporus for potential 7-mer binding sites, using the 44 most highly up-regulated transcripts in the Zfs1 deficient strains that contained at least one 7-mer in C. albicans.
Molecular phylogenetic analysis
Protein sequence relatedness for the 64-amino acid TZF domains from 14 members of the CTG clade was calculated using the neighbor-joining method as implemented in MEGA5.1 (Tamura et al., 2011), using the Dayhoff and bootstrap algorithms available in the program. The tree is drawn to scale, with branch lengths represented in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Phenotypic characterization of zfs1Δ/Δ mutants
To assess hyphal formation in liquid medium, strains were grown planktonically at 37°C under three types of filament-inducing conditions as described in Nobile et al., (2012): (i) Roswell Park Memorial Institute medium (RPMI) medium for 90 min, (ii) Spider medium for 3 h and (iii) YPD + 10% serum for 2 h. Strains were inoculated from saturated overnight YPD cultures into the corresponding filament-inducing medium at an OD600 = 0.2. Two hundred cells from each medium were counted and analyzed for hyphal formation by light microscopy.
To assess hyphal formation on solid medium, strains were grown on plates at 37°C under four types of filament-inducing conditions as described in Homann et al. (2009): (i) YPD + 10% serum, (ii) Lee’s (pH 4.5) medium, (iii) Spider medium and (iv) blood agar. Colony morphology phenotypes were observed visually for filamentation daily for 7 days after plating. For comparison, strains were also grown on YPD medium under the same conditions.
To assess biofilm formation, strains were grown as biofilms in Spider medium on the bottom of 6-well polystyrene plates under standard in vitro conditions (Nobile et al., 2012). Biofilms were visualized by confocal scanning laser microscopy (CSLM) as described in Nobile et al. (2012). For CSLM, biofilms were stained with 50 µg ml−1 of concanavalin AAlexa Fluor 594 conjugate (conA-594) (Molecular Probes) in the dark for 1 h with 200 r.p.m. agitation at 37°C. CSLM was performed in the Nikon Imaging Center at UCSF with a Nikon Eclipse C1si upright spectral imaging confocal microscope using a 40 ×/0.80 W Nikon objective. For conA-594 visualization, a 561 nm laser line was used. Images were acquired using Nikon EZ-C1 Version 3.80 software, and assembled into top views and maximum intensity Z-stack projections (side views) using Nikon NIS Elements Version 3.00 software.
Growth curves
Growth curves were performed by inoculating cells from an overnight culture grown in YPD at 30°C to a starting OD600 = 0.01 in 100 µl YPD medium as described in Singh-Babak et al. (2012). Growth curves were performed in flat-bottom 96-well microtiter plates (Costar) and grown in a Tecan Infinite M1000 reader (Tecan Systems) at 30°C with 408 r.p.m. orbital shaking. Optical density (OD595) measurements were taken every 15 min for 24 h.
Flocculation assay
Flocculation was performed as described previously (Soares and Mota, 1997; Wells et al., 2012). Briefly, 5 ml of logarithmically growing cultures was centrifuged at 4500 × g for 5 min at 4°C. Cells were then washed twice in 250 mM EDTA, once in 250 mM NaCl (pH 2) and finally in 250 mM NaCl (pH 4.5). Washed cells were resuspended to a final concentration of 1 × 108 cells ml−1 in 25 ml of 250 mM NaCl (pH 4.5) and placed into a 25 ml graduated cylinder. The cell suspension was adjusted to 4 mM CaCl2 with 100 mM CaCl2 (pH 4.5) and inverted 18 times. At defined intervals, 100 µl of cell suspension was removed from a fixed position in the graduated cylinder and diluted with 900 µl of water, and absorbance was then immediately measured at optical density 600 nm. All values were normalized to 100% of cells in suspension at time point 0.
Expression and purification of recombinant Zfs1
Full-length C. albicans ZFS1 (orf19.5334) was cloned using the NotI and BamHI sites of a modified pMAL-c5x vector (New England Biolabs), as described in Moon et al. (2010), and was expressed as a fusion protein with maltose-binding protein linked to the N-terminus of Zfs1 (Zfs1:MBP) by a three alanine linker. The recombinant fusion protein was expressed in BL21 (DE3) cells after induction with 0.3 mM IPTG for 16 h at 20°C. Cells were lysed by sonication and cleared by centrifugation for 35 min at 35 000 × g. The resulting supernatants were incubated for 1 h at 4°C with amylose resin (New England Biolabs). Zfs1:MBP was eluted with 40 mM maltose. Fusion protein-containing fractions were subsequently applied to a Superdex 200 size-exclusion column (GE Healthcare), followed by further purification on a HiTrap Q (GE Healthcare) anion exchange column. Fractions containing purified Zfs1:MBP were pooled and concentrated using a 30 000 molecular weight cut-off (MWCO) Vivaspin 20 centrifugal filtering device (GE Healthcare), and stored at −80°C after flash freezing in liquid nitrogen. Coomassie blue staining of Sodium Dodecyl Sulfate (SDS) polyacrylamide gels indicated a purity of this final preparation of approximately 90–95%.
Determination of RNA binding affinity by fluorescence anisotropy
RNA probes were synthesized and purified, and 2′-hydroxyl groups deprotected by Dharmacon. Fluorescence anisotropy experiments were carried out using 5′-fluorescein-labeled RNA probes, as described previously (Wilson et al., 2001a,b; Blackshear et al., 2003). Briefly, lyophilized RNA probes were resuspended in 10 mM Tris, pH 8. Assays were carried out at 25°C in a final volume of 100 µl in 10 mM Tris (pH 8.0), 50 mM KCl, 2 mM DTT, 0.2 mg ml−1 heparin, 0.1 mg ml −1 bovine serum albumin and 5 µM ZnCl2. RNA binding affinity of the purified Zfs1:MBP fusion protein was measured using the Beacon 2000 variable temperature fluorescence polarization system (Panvera) containing fluorescein excitation (λex = 490 nm) and emission (λem = 535 nm) filters. Anisotropy was measured over 1 min and averaged, which previously was determined to be sufficient for binding equilibrium to be reached (data not shown) (Wilson et al., 2001b). In all experiments described, total fluorescence emission was measured to verify that protein binding did not alter the fluorescence quantum yield of fluorescein-labeled RNA substrates (data not shown) (Blackshear et al., 2003).
Total measured anisotropy (At) was measured over a range of protein (P) concentrations. Binding constants were calculated by the nonlinear regression algorithm in PRISM, version 6.0 (GraphPad Software) using Eq. 1 (Wilson et al., 2001a).
| (1) |
K represents the apparent equilibrium constant (K; K = 1/Kd); AR and APR are the intrinsic anisotropy values of free RNA and the protein-fluorescein-labeled RNA complex respectively. Data are plotted as Δanisotropy, where the anisotropy of the probe alone was subtracted from the total anisotropy measured at each protein concentration.
RNA electrophoretic mobility shift assay
Gel shift analysis was performed as described previously (Kedar et al., 2012) with the following modification: 0.01, 0.1 or 1.0 µg of purified Zfs1:MBP was incubated with 0.6 ng of a 5′-biotin-labeled (Invitrogen) probe derived from the 3′UTR of mouse TNF alpha mRNA (bp 1309–1332 from GenBank accession number X02611).
RIP analysis
Zfs1:Myc and a no tag (SN250) strain were grown overnight in YPD media. Cultures were then diluted to OD600 ∼ 0.1 in 200 ml of YPD media and grown at 30°C for 5 h. Cells were harvested by centrifugation and lysed as described previously (Coleman et al., 2014). Ten percent of the clarified lysate was removed for an input sample and total RNA was purified using the RiboPure Yeast RNA Purification kit following the manufacturer’s protocol. To immunoprecipitate Zfs1:Myc, extracts were incubated with Myc-agarose (9E10, Santa Cruz Biotechnology) for 2 h at 4°C. The agarose was washed four times in 25 mM HEPES-KOH (pH 7.5), 150 mM KCl2 and 2 mM MgCl2. Immunoprecipitated RNA was eluted using the RiboPure Yeast RNA Purification kit following the manufacturer’s protocol. cDNAs from input and immunoprecipitated RNA samples were made using the iScript cDNA synthesis kit (Biorad) and subjected to real-time RT-PCR analysis with primers spanning the indicated transcripts. Fold enrichment was calculated as described previously (Foureau et al., 2014). The ΔCt normalized immunoprecipitated sample was calculated by (Ct [RIP] – (Ct [Input] – Log2 (Input Dilution Factor)), where input dilution factor = fraction of the input RNA saved. To calculate the % Input for each RIP fraction, the following equation was used: % Input = 100 × 2−ΔCt[normalized RIP]. To adjust the normalized RIP fraction Ct value against the normalized background [no tag strain (NS)] fraction Ct value (first ΔΔCt), we used ΔΔCt [RIP/NS] = ΔCt [normalized RIP] – ΔCt [normalized NS]. To calculate fold enrichment above the no tag strain background, the following equation was used: Fold Enrichment = 2−ΔΔCt[RIP/NS]
Acknowledgements
We thank the NIH Intramural Sequencing Center for performing the mRNA-Seq analysis and Tom Randall for assistance with tree generation. We are grateful for the availability of the Nikon Imaging Center (NIC) at UCSF, where confocal scanning laser microscopy images were acquired. We also thank Jessica Williams and Jim Westmoreland for critical comments on the manuscript, and Alexander Johnson for providing the wild-type and zfs1Δ/Δ mutant strains. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. CJN was supported by NIH grant R00AI100896; NH and CJN were supported by NIH grant R01AI083311 to Alexander Johnson; and GMW and BEZ were supported by NIH grant R01CA102428.
References
- Bai C, Elledge SJ. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- Bart-Delabesse E, Sarfati J, Debeaupuis JP, van Leeuwen W, van Belkum A, Bretagne S, Latge JP. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates. J Clin Microbiol. 2001;39:2683–2686. doi: 10.1128/JCM.39.7.2683-2686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X, Zhou P. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J Biol Chem. 2003;278:19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, d’Enfert C. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol. 2011;80:995–1013. doi: 10.1111/j.1365-2958.2011.07626.x. [DOI] [PubMed] [Google Scholar]
- Botterel F, Desterke C, Costa C, Bretagne S. Analysis of microsatellite markers of Candida albicans used for rapid typing. J Clin Microbiol. 2001;39:4076–4081. doi: 10.1128/JCM.39.11.4076-4081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- Brewer BY, Ballin JD, Fialcowitz-White EJ, Blackshear PJ, Wilson GM. Substrate dependence of conformational changes in the RNA-binding domain of tristetraprolin assessed by fluorescence spectroscopy of tryptophan mutants. Biochemistry. 2006;45:13807–13817. doi: 10.1021/bi061320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA, Blackshear PJ. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta. 2013;1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HG, Bhat SK, Murray LJ, McManus DT, O’Neill OM, Gavin AT, Johnston BT. Symptoms and endoscopic features at Barrett’s esophagus diagnosis: implications for neoplastic progression risk. Am J Gastroenterol. 2014;109:527–534. doi: 10.1038/ajg.2014.10. [DOI] [PubMed] [Google Scholar]
- Cuthbertson BJ, Liao Y, Birnbaumer L, Blackshear PJ. Characterization of zfs1 as an mRNA-binding and -destabilizing protein in Schizosaccharomyces pombe . J Biol Chem. 2008;283:2586–2594. doi: 10.1074/jbc.M707154200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, et al. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foureau E, Clastre M, Montoya EJ, Besseau S, Oudin A, Glevarec G, et al. Subcellular localization of the histidine kinase receptors Sln1p, Nik1p and Chk1p in the yeast CTG clade species Candida guilliermondii . Fungal Genet Biol. 2014;65:25–36. doi: 10.1016/j.fgb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Gomez-Raja J, Andaluz E, Magee B, Calderone R, Larriba G. A single SNP, G929T (Gly310Val), determines the presence of a functional and a non-functional allele of HIS4 in Candida albicans SC5314: detection of the non-functional allele in laboratory strains. Fungal Genet Biol. 2008;45:527–541. doi: 10.1016/j.fgb.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Umbach DM, Vincent Jordan N, Abell AN, Johnson GL, Li L. Efficiently identifying genome-wide changes with next-generation sequencing data. Nucleic Acids Res. 2011;39:e130. doi: 10.1093/nar/gkr592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Sugimoto A, Yamamoto M. Schizosaccharomyces pombe zfs1+ encoding a zinc-finger protein functions in the mating pheromone recognition pathway. Mol Biol Cell. 1995;6:1185–1195. doi: 10.1091/mbc.6.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedar VP, Zucconi BE, Wilson GM, Blackshear PJ. Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins. J Biol Chem. 2012;287:5459–5471. doi: 10.1074/jbc.M111.312652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Carrick DM, Blackshear PJ. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J Biol Chem. 2005;280:34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, et al. Regulatory networks affected by iron availability in Candida albicans . Mol Microbiol. 2004;53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- L’Ollivier C, Labruere C, Jebrane A, Bougnoux ME, d’Enfert C, Bonnin A, Dalle F. Using a Multi-Locus Microsatellite Typing method improved phylogenetic distribution of Candida albicans isolates but failed to demonstrate association of some genotype with the commensal or clinical origin of the isolates. Infect Genet Evol. 2012;12:1949–1957. doi: 10.1016/j.meegid.2012.07.025. [DOI] [PubMed] [Google Scholar]
- Ma Q, Herschman HR. The yeast homologue YTIS11, of the mammalian TIS11 gene family is a non-essential, glucose repressible gene. Oncogene. 1995;10:487–494. [PubMed] [Google Scholar]
- McCourtie J, Douglas LJ. Relationship between cell surface composition, adherence, and virulence of Candida albicans . Infect Immun. 1984;45:6–12. doi: 10.1128/iai.45.1.6-12.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus BA, Coleman DC. Molecular epidemiology, phylogeny and evolution of Candida albicans . Infect Genet Evol. 2014;21:166–178. doi: 10.1016/j.meegid.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LG, Hajjeh RA, Edwards JE., Jr Estimating the cost of nosocomial candidemia in the United States. Clin Infect Dis. 2001;32:1110. doi: 10.1086/319613. [DOI] [PubMed] [Google Scholar]
- Moon AF, Mueller GA, Zhong X, Pedersen LC. A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010;19:901–913. doi: 10.1002/pro.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro FJ, Nurse P. A systematic screen reveals new elements acting at the G2/M cell cycle control. Genome Biol. 2012;13:R36. doi: 10.1186/gb-2012-13-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, et al. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans . Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Ricarte F, Menjivar R, Chhun S, Soreta T, Oliveira L, Hsueh T, et al. A genome-wide immunodetection screen in S. cerevisiae uncovers novel genes involved in lysosomal vacuole function and morphology. PLoS ONE. 2011;6:e23696. doi: 10.1371/journal.pone.0023696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans . Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RM, Dayhoff MO. Protein and nucleic acid sequence data and phylogeny. Science. 1979;205:1038–1039. doi: 10.1126/science.205.4410.1038. [DOI] [PubMed] [Google Scholar]
- Singh-Babak SD, Babak T, Diezmann S, Hill JA, Xie JL, Chen YL, et al. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata . PLoS Pathog. 2012;8:e1002718. doi: 10.1371/journal.ppat.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares EV, Mota M. Quantification of yeast flocculation. J Inst Brew. 1997;103:93–98. [Google Scholar]
- Stajich JE, Berbee ML, Blackwell M, Hibbett DS, James TY, Spatafora JW, Taylor JW. The fungi. Curr Biol. 2009;19:R840–R845. doi: 10.1016/j.cub.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, Lai WS, Taylor GA, Blackshear PJ. Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene. 1996;174:225–233. doi: 10.1016/0378-1119(96)00084-4. [DOI] [PubMed] [Google Scholar]
- Vergara SV, Puig S, Thiele DJ. Early recruitment of AU-rich element-containing mRNAs determines their cytosolic fate during iron deficiency. Mol Cell Biol. 2011;31:417–429. doi: 10.1128/MCB.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells ML, Huang W, Li L, Gerrish KE, Fargo DC, Ozsolak F, Blackshear PJ. Posttranscriptional regulation of cell-cell interaction protein-encoding transcripts by Zfs1p in Schizosaccharomyces pombe . Mol Cell Biol. 2012;32:4206–4214. doi: 10.1128/MCB.00325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Sutphen K, Bolikal S, Chuang KY, Brewer G. Thermodynamics and kinetics of Hsp70 association with A + U-rich mRNA-destabilizing sequences. J Biol Chem. 2001a;276:44450–44456. doi: 10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Sutphen K, Chuang K, Brewer G. Folding of A + U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J Biol Chem. 2001b;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]