Abstract
Background
Pedunculate oak, Quercus robur is an abundant forest tree species that hosts a large and diverse community of beneficial ectomycorrhizal fungi (EMFs), whereby ectomycorrhiza (EM) formation is stimulated by mycorrhiza helper bacteria such as Streptomyces sp. AcH 505. Oaks typically grow rhythmically, with alternating root flushes (RFs) and shoot flushes (SFs). We explored the poorly understood mechanisms by which oaks integrate signals induced by their beneficial microbes and endogenous rhythmic growth at the level of gene expression. To this end, we compared transcript profiles of oak microcuttings at RF and SF during interactions with AcH 505 alone and in combination with the basidiomycetous EMF Piloderma croceum.
Results
The local root and distal leaf responses to the microorganisms differed substantially. More genes involved in the recognition of bacteria and fungi, defence and cell wall remodelling related transcription factors (TFs) were differentially expressed in the roots than in the leaves of oaks. In addition, interaction with AcH 505 and P. croceum affected the expression of a higher number of genes during SF than during RF, including AcH 505 elicited defence response, which was attenuated by co-inoculation with P. croceum in the roots during SF. Genes encoding leucine-rich receptor-like kinases (LRR-RLKs) and proteins (LRR-RLPs), LRR containing defence response regulators, TFs from bZIP, ERF and WRKY families, xyloglucan cell wall transglycolases/hydrolases and exordium proteins were differentially expressed in both roots and leaves of plants treated with AcH 505. Only few genes, including specific RLKs and TFs, were induced in both AcH 505 and co-inoculation treatments.
Conclusion
Treatment with AcH 505 induces and maintains the expression levels of signalling genes encoding candidate receptor protein kinases and TFs and leads to differential expression of cell wall modification related genes in pedunculate oak microcuttings. Local gene expression response to AcH 505 alone and in combination with P. croceum are more pronounced when roots are in resting stages, possibly due to the fact that non growing roots re-direct their activity towards plant defence rather than growth.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1856-y) contains supplementary material, which is available to authorized users.
Keywords: Plant growth promoting rhizobacteria (PGPR), Defence, Disease resistance, Mycorrhiza
Background
Soil microbial communities influence numerous physiological processes and traits of plants, including: seed germination, seedling vigour, growth and development, nutrition and the progression of various diseases [reviewed in 1]. Mutualistic fungi form close associations with plants that provide multiple benefits to both sets of organisms. Most temperate forest trees develop a mutualistic root symbiosis, ectomycorrhiza (EM), with fungi. This enhances the tree nutrient acquisition and the fungus ability to extend mycelia through the soil and form fruiting bodies [2]. Various other beneficial microorganisms are also frequently associated with EM, including mycorrhiza helper bacteria (MHBs) [3]. Production of bacterial secondary metabolites such as auxofuran [4] stimulate mycelial growth, but mycorrhiza formation can also be promoted by the reduction of soil-mediated stress [5, 6] and by augmentation of plant-fungus contacts by stimulation of lateral root formation [7]. Helper activities can also improve mycorrhizal functioning, for instance by mobilising nutrients from soil minerals [8] and protecting plants against attack by root pathogens [9]. Although the importance of EM-associated bacteria for the symbiosis is established, it is unclear how plants coordinate their gene expression responses to the MHB in the presence and absence of the EMF.
Like many tropical and some temperate trees, oaks display an endogenous rhythmic growth pattern with alternating phases of development and rest throughout their vegetative growth periods [10]. The endogenic character of rhythmic growth is reflected by the regular period of alternating shoot flushes (SF) and root flushes (RF) under uniform and constant environmental conditions [11–13]. It has been suggested that the rhythmic development is a consequence of carbon partitioning [14–17]. Herrmann, et al. [18] developed a microcosm system in which microcuttings of the pedunculate oak Quercus robur clone DF159 express these typical alternating flushes and using the same system, Angay, et al. [19] showed that concentrations of high nonstructural carbohydrates in roots are high during RF and low during SF.
Rhythmic growth of oak trees affects their interactions with associated microorganisms. For instance the distribution pattern on the mother roots and the frequency of mycorrhizal root tips are affected by the endogenous rhythmic growth of Q. robur [10]. In addition, infestation of roots by the oomycete pathogen Phytophthora quercina is enhanced during RF, when roots are growing rapidly and the plant below-ground carbon allocation is highest [19], and colonisation of leaves by oak powdery mildew is enhanced during SF, when the leaves are growing most rapidly [20]. Oaks are a group of major broadleaf forest trees in Europe that are key components of complex networks of biotrophic interactions, involving relationships with endophytic [21], pathogenic [22] and EM [23] fungi, as well as bacteria that stimulate EM formation [24]. Since the impact of rhythmic growth on biotic interactions of the oak is so evident, it should be investigated if it affects the gene expression response of the oak to these interacting organisms.
Previously, we showed that co-inoculation of oak microcuttings with the mycorrhiza helper bacterium Streptomyces sp. strain AcH 505 (hereafter AcH 505) led to an increase in the number of Piloderma croceum-mycorrhizal plants [25]. Furthermore, AcH 505 induced a systemic defence response against Microsphaera alphitoides in pedunculate oak leaves, involving both jasmonic acid (JA)/ethylene (ET) and salicylic acid (SA)-dependent signalling [26]. However, the cited studies did not investigate variations in transcriptomic responses to AcH 505 during RF and SF growth stages, effects of AcH 505-treatment in roots, or its interactions with mycorrhizal fungi, which are crucial elements of a MHB functional role in the oak system.
EMFs, such as P. croceum are essential for optimal development of pedunculate oak microcuttings [23, 27]. Extensive re-programming of the oak transcriptome has been detected both during pre-symbiotic development and in mature symbiotic EM with P. croceum [28–30]. For instance, EM formation with P. croceum leads to defence suppression in oak, embodied by low abundance of defence-related transcripts [30]. But the influence of dual presence of AcH 505 and P. croceum on the oak was not investigated.
When microorganisms interact with plants, perception processes play a crucial role. Plants perceive microorganisms by sensing microbe-associated molecular patterns [MAMPs; 31]. MAMP receptors of plants are encoded by R genes including transmembrane receptor-like kinases (RLKs), receptor-like proteins (RLPs) and leucine rich repeat-nucleotide binding (LRR-NB) proteins [32] and they can recognize both pathogenic [33] and beneficial microorganisms [31]. Transcription factors (TFs) of the APETALA 2/ethylene-responsive family, in particular ethylene-responsive element binding factors (ERFs) play a major role in integrating the response to biotic interactions and are implicated in MAMP-induced systemic resistance signalling [34]. Seven ERF contigs showed up-regulation in P. croceum/oak EM [30], but it has not been investigated how AcH 505 treatment affects their transcription.
Based on our analysis of systemic defence response in leaves to AcH 505 [26] and on the defence suppressing effect of P. croceum on oaks [30] we questioned i) how the oak coordinates its gene expression responses to MHB in the absence and presence of the EMF, ii) how the gene expression response of the oak to the interacting organisms is affected by the stage of growth, iii) if AcH 505 affects the P. croceum attenuated defence related gene expression in oak or vice versa, and iv) if AcH 505 treatment affects the transcription of ERFs. This work was based on two hypotheses. Our first hypothesis states that AcH 505 induces, while co-inoculation with P. croceum attenuates the defence responses in oak. Our second hypothesis states that local molecular responses to the organisms interacting with their roots are stronger during SF, expressed as differential representation of R genes, hormonal signalling genes and cell wall proteins. The second hypothesis is based on the observations of Angay, et al. [19] that oak roots express a higher level of root colonisation by P. quercina in RF, and also, on the general literature on MAMP perception, hormonal signalling and cell wall modifications occurring during plant-microbe interactions [e.g. 33, 34]. To address these hypotheses, we investigated transcriptional alterations in oak during the colonisation by AcH 505 and the co-colonisation with P. croceum, focussing on the responses of roots and leaves during RF and SF. The gene expression data demonstrates that the transcriptional response of the oak is more active during SF, the period when the roots are not growing, that a defence response is induced by the mycorrhization helper bacterium but attenuated by the EMF, and that several candidate genes related to MAMP recognition processes and control of transcription are differentially expressed (DE) upon these treatments.
Results
Oak microcuttings were successfully inoculated with AcH 505 (Fig. 1). Three replicate RNA-Seq datasets were generated for microcutting lateral roots and leaves, during both RF and SF, following no inoculation, inoculation with AcH 505 and with both AcH 505 and P. croceum (n = 36; Additional files 1 and 2). For differential gene expression analysis, pairwise comparisons of datasets from plants inoculated with AcH 505 versus non-inoculated plants [differentially expressed contigs (DECs) in AcH 505 inoculated plants] and plants inoculated with AcH 505 and P. croceum versus non-inoculated plants [DECs in co-inoculated plants] were performed for roots and leaves at RF and SF respectively. The validity of the differential expression analyses was confirmed by qRT-PCR analysis (Additional file 3). Numbers of DECs among the samples are visualised in the Venn diagrams shown in Fig. 2 and tabulated in Additional file 4. AcH 505 treatment induced more DECs during SF than during RF in both roots and leaves. A suppressive effect by AcH 505-P. croceum co-inoculation on the numbers of DECs was evident during SF. Proportions of contigs regulated in AcH 505 inoculated plants which were also regulated when subjected to co-inoculation with P. croceum, were 33 in roots and 23 % in leaves during RF, and 5 and 3 % during SF.
Fig. 1.

Pedunculate oak microcuttings with interacting microorganisms. Streptomyces sp. AcH 505 on soil particles in the microcosm (a) Piloderma croceum – oak ectomycorrhizas (b) and a pedunculate oak Quercus robur microcutting (c)
Fig. 2.
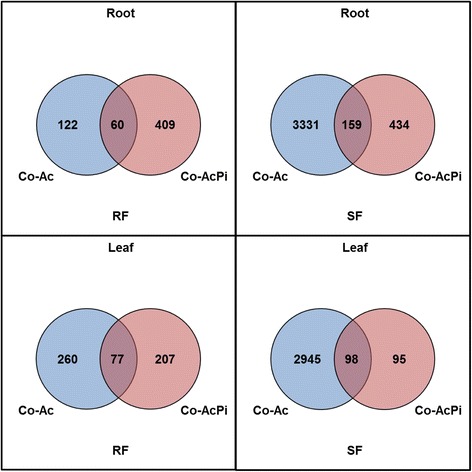
Venn-diagrams illustrating numbers of DECs (Benjamini-Hochberg adjusted, p ≤ 0.01). The comparisons depicted are for the following pairs of roots and leaves of microcuttings during RF and SF: Control versus AcH 505 inoculated (Co-Ac) and Control- versus AcH 505- and P. croceum-inoculated (Co-AcPi)
To obtain a comprehensive view of differential gene expression, we conducted functional annotation enrichment analyses with GOseq. The 10 most significantly enriched terms in Protein family (Pfam) and Gene Ontology (GO) categories are shown in Additional file 5. Representative enriched Pfam terms are shown in Fig. 3, enriched plant defence-related GO terms in roots in Fig. 4, and representative DECs for the Pfam and GO terms, as well as plant defence-related DECs in roots, are listed in Additional file 6.
Fig. 3.
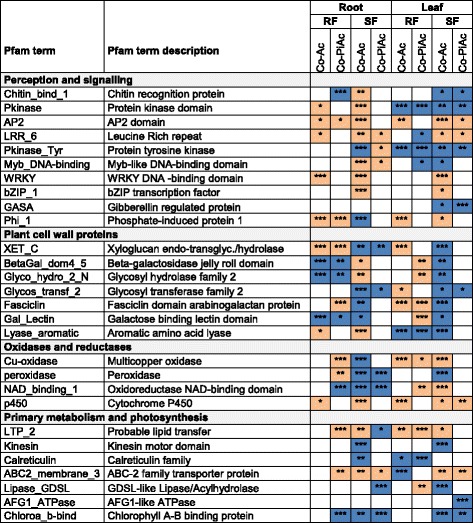
Protein family (Pfam) terms enriched for up- and down-regulated contigs, in roots and leaves harvested from plants during RF and SF, treated with AcH 505 (Co-Ac) and both microorganisms (Co-AcPi). Putative function categories “perception and signalling”, “plant cell wall proteins”, “oxidases and reductases” and “primary metabolism and photosynthesis” are given. Orange colour indicates up-regulated and blue down-regulated enriched Pfam terms. Significance levels are marked by asterisks in the boxes (*** :p ≤ 0.001; **: 0.001 < p ≤ 0.01; *: 0.01 < p ≤ 0.05). The p-value cut-off was set at p ≤ 0.05
Fig. 4.
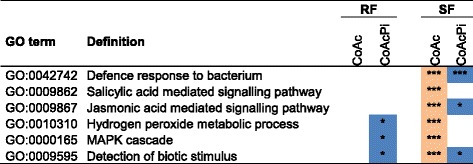
Enriched plant defence-related GO terms in pedunculate oak roots, showing terms enriched for up-regulated contigs after AcH 505 inoculation specifically during SF and partially depleted in co-inoculated plants. GO enrichment analysis was implemented by GOseq. Orange colour indicates up-regulated and blue down-regulated enriched Pfam terms. Significance levels are marked by asterisks in the boxes. (*** :p ≤ 0.001; **: 0.001 < p ≤ 0.01; *: 0.01 < p ≤ 0.05). The p-value cut-off was set at p ≤ 0.05
Pfam enrichment analysis revealed significant changes in Pfam terms related to perception and signalling, plant cell wall proteins, oxidases and reductases, as well as primary metabolism and photosynthesis in both AcH 505- and co-inoculated plants (Fig. 3; Additional file 5). In roots at RF, plant cell wall protein related Pfam terms were strongly enriched. For instance, Xyloglucan endo-transglycosylase (XET) was enriched in up-regulated contigs (EUC) in both treatments and Fasciclin domain arabinogalactan protein (FDA) in co-inoculated plants, with other EUC including Phosphate-induced protein 1 conserved region (PIP1) and Probable lipid transfer (LTP). The Pfam term EUC was associated with increased abundance of several predicted XET, PIP1, FDA and LTP transcripts (Additional file 6). The terms which were enriched in down-regulated contigs (EDC) included Beta-galactosidase and Galactose binding lectin domain in AcH 505 treatment, but photosynthesis and plant defence related terms in co-inoculation, with Pfam terms Chlorophyll A-B binding protein (CHLBP) and Chitin recognition protein. In roots at SF under AcH 505 treatment, the most significant Pfam terms EUC were related to stress associated transcriptional regulation, such as Bzip and WRKY DNA-binding domain, the latter with seven up-regulated candidate WRKY contigs (Additional file 6). AcH 505 treatment led to Pfam terms EDC of the types Multicopper oxidase and Glycosyl transferase family 2, and co-inoculation the term CHLBP with 24 down-regulated candidate CHLBP contigs (Additional file 6), as well as the development and defence response related term GDSL-like Lipase/Acylhydrolase (GDSL). In leaves at RF, the terms EUC included PIP1 and XET with associated contigs (Additional file 6) in AcH 505 treatment, and LTP and Galactose binding lectin domain in co-inoculation. The terms EDC included Protein kinase domain and Calreticulin in both treatments and ABC-2 transporter in AcH 505 treatment. In leaves at SF, the terms EUC included Multicopper oxidase in AcH 505 treatment, and GDSL in co-inoculation. In contrast, the terms EDC included Kinesin motor domain and GDSL in AcH 505 treatment, and Gibberellin regulated protein and AFG1-like ATPase in co-inoculation.
AcH 505 inoculation led to GO term enrichment which corroborated qualitatively with Pfam term results (Additional file 5). For instance in roots at RF, plant cell wall GO term Xyloglucan:xyloglucosyl transferase activity was EUC in both treatments in corroboration with the Pfam term XET, and Lipid transport in co-inoculation, in line with the Pfam term LTP. Plant defence associated GO terms were prominently EUC upon AcH 505 treatment in roots, and when the oaks were co-inoculated with the EMF, these GO-terms were EDC or non-significant (Fig. 4). Among the contigs which were up-regulated upon AcH 505 but not by co-inoculation, we detected disease resistance, protein kinase, TF, and chitinase contigs (Additional file 6), indicating that suppression of plant defence by P. croceum [30] overrules the defence-stimulating effect of AcH 505 in roots.
To address the AcH 505 driven gene expression in more detail, we searched for those individual contigs that were regulated as well in AcH 505 as in co-inoculation (Table 1). We reasoned that those genes regulated in AcH 505 but not influenced by additional presence of P. croceum, may be part of the core interactome gene set of AcH 505 on the oak. In the roots of plants during RF, these core contigs were related to plant growth (XET) and signalling (Kinase-like protein). During SF, the core contigs were related to transcriptional regulation and redox reactions (Chromatin binding protein, Peroxidase), and the down-regulated contigs included photosynthesis related PS II binding protein LHCB1.5. In the leaves of the microcuttings, based on the Arabidopsis thaliana orthologs, the contigs with corresponding patterns identified during RF were related to cell wall biosynthesis (Cellulose synthase, Fasciclin-like arabinogalactan-protein), lipid transport or signalling (Lipid transfer protein), and transport (PHO1 phosphate exporter), whilst during SF the core contigs were involved in defence and growth-related signalling (Calcium-binding protein, LRR nucleotide binding sequence [NBS] resistance protein, anthocyanidin synthase).
Table 1.
Contigs regulated in AcH 505 inoculated plants which were not altered when subjected to co-inoculation with P. croceum in roots and leaves and during RF and SF
| Co-Ac | Co-PiAc | ||||
|---|---|---|---|---|---|
| Contig | Log2 fold change | FDR | Log2 fold change | FDR | Sequence description |
| Root – RF | |||||
| comp34428_c0_seq2 | 8,8 | 2,0E-07 | 7,7 | 1,3E-04 | Ice binding |
| comp43120_c0_seq11 | 2,6 | 7,4E-07 | 2,2 | 6,6E-17 | Xyloglucan endotransgl/hydrolase |
| comp36512_c0_seq1 | 5,9 | 1,1E-06 | 6,0 | 5,3E-08 | Nucleolar complex protein |
| comp32163_c0_seq1 | 2,2 | 1,3E-05 | 2,9 | 8,3E-09 | Clavaminate synthase |
| comp43753_c1_seq2 | 8,6 | 9,0E-05 | 8,6 | 3,9E-08 | Kinase-like protein |
| comp39841_c1_seq1 | 1,1 | 6,0E-05 | 1,1 | 2,0E-06 | Exordium-like protein |
| comp37819_c1_seq5 | 7,9 | 1,5E-03 | 8,5 | 3,6E-10 | o-linked c transferase |
| comp43557_c1_seq2 | 8,4 | 7,0E-03 | 8,6 | 2,2E-10 | tRNA-dihydrouridine synthase |
| Root – SF | |||||
| comp29599_c0_seq1 | −1,3 | 6,6E-19 | −1,9 | 9,9E-03 | Gibberellin-regulated protein |
| comp41819_c4_seq2 | −8,5 | 1,3E-10 | −8,7 | 4,5E-07 | F-box family protein |
| comp43426_c1_seq15 | −7,2 | 4,8E-07 | −7,4 | 1,9E-04 | Sugar transporter |
| comp37704_c0_seq2 | −1,0 | 3,1E-04 | −3,9 | 5,5E-44 | PS II binding protein LHCB1.5 |
| comp42662_c0_seq8 | 1,6 | 1,1E-99 | 1,1 | 3,1E-05 | Cytochrome p450 |
| comp19461_c0_seq1 | 1,2 | 7,0E-54 | 1,1 | 1,4E-04 | Peroxidase 10 |
| comp35114_c0_seq1 | 1,1 | 1,4E-45 | 1,4 | 5,4E-04 | Zinc finger protein |
| comp19664_c0_seq1 | 0,8 | 2,2E-11 | 1,0 | 2,3E-03 | Thaumatin |
| comp42100_c0_seq10 | 7,2 | 9,2E-06 | 6,9 | 7,8E-04 | Chromatin binding |
| Leaf – RF | |||||
| comp36279_c0_seq4 | −9,0 | 1,1E-09 | −8,9 | 1,9E-09 | GTP cyclohydrolase |
| comp23339_c0_seq1 | −1,5 | 1,6E-06 | −1,8 | 7,6E-10 | Phosphoglycerate dehydrogenase |
| comp40475_c2_seq3 | −1,1 | 8,2E-04 | −0,9 | 3,0E-03 | Leucine-rich repeat receptor |
| comp28232_c0_seq1 | 2,3 | 4,6E-04 | 2,0 | 5,4E-04 | Fasciclin-like arabinogalactan-protein |
| comp30075_c0_seq1 | 1,8 | 4,8E-03 | 2,2 | 1,4E-05 | Lipid transfer protein |
| comp42008_c3_seq5 | 3,7 | 3,5E-06 | 3,1 | 2,4E-03 | Calcium-binding protein |
| comp39615_c0_seq1 | 2,2 | 3,9E-06 | 1,8 | 9,3E-04 | Multicopper oxidase |
| comp42669_c0_seq2 | 5,1 | 1,2E-05 | 3,8 | 6,2E-03 | Cellulose synthase |
| comp42379_c0_seq7 | 4,3 | 9,0E-03 | 4,4 | 2,1E-03 | Pho1-like protein |
| Leaf – SF | |||||
| comp30731_c0_seq2 | −0,7 | 1,9E-20 | −0,9 | 1,3E-03 | Cbl-interacting serine threonine-protein |
| comp30800_c0_seq1 | −2,7 | 1,8E-06 | −2,2 | 3,6E-03 | Adenylyl-sulfate reductase |
| comp39235_c1_seq5 | −6,7 | 1,8E-04 | −6,7 | 5,8E-03 | Chaperone protein |
| comp21202_c0_seq1 | 3,4 | 3,4E-85 | 1,4 | 1,7E-04 | Anthocyanidin synthase |
| comp42008_c3_seq5 | 2,5 | 1,6E-41 | 2,6 | 6,0E-04 | Calcium-binding protein |
| comp42290_c0_seq1 | 1,2 | 3,6E-27 | 1,8 | 9,2E-10 | Lipoxygenase |
| comp42379_c0_seq7 | 8,6 | 5,4E-11 | 9,0 | 1,2E-03 | Pho1-like protein |
| comp43826_c1_seq6 | 8,8 | 4,1E-10 | 7,9 | 1,1E-05 | LRR-NB-ARC domain protein |
Significant differential expression was determined by edgeR with a threshold Benjamini-Hochberg adjusted p-value of 0.01, indicated by "FDR"
Abbreviations: endotransgl, endotransglycosidase; PSII, photosystem II
Due to the Pfam term enrichment in both treatments, both stages of growth, and tissues, the Leucine Rich Repeat (LRR) and Ethylene response transcription factor (AP2/ERF)-associated DECs were treated in more detail and their homology to orthologous transcripts in A. thaliana was estimated by blastx analysis (Additional file 6). Most of the predicted LRR contigs encoded members of the extensive families of LRR receptor-like kinases (LRR-RLKs) and receptor-like proteins (LRR-RLPs) with roles in A. thaliana ranging from recognition of microorganisms to signalling in growth and differentiation. The dominance of SF vs. RF in the numbers of DECs was particularly strong for the predicted LRR containing defence response regulators. From the 34 DECs of this type, 31 occurred at SF but only 3 at RF (Additional file 6). These DECs comprised candidate leucine-rich repeat-nucleotide binding site (LRR-NBS) contigs, which act as R genes in A. thaliana. Predicted functions of other identified LRR-encoding DECs are related to auxin responses, microtubule cytoskeleton, cell wall composition and terpenoid biosynthesis. Among these signalling devices and TFs, which are most important for the AcH 505 interaction, contigs regulated in both AcH 505- and co-inoculations were detected. From LRR-receptor protein kinase encoding contigs, a contig (homologous to A. thaliana AT3G47110) was up-regulated in leaves, and another (AT3G14840), was down-regulated in leaves at RF and SF by both treatments. Contig encoding LRR-NB-ARC domain protein (AT3G14460) was up-regulated in both roots and leaves at SF. From TFs, AP2/ERF contig homologous to A. thaliana SHN2 (AT5G11190) was up-regulated in roots and leaves at RF by both treatments, and by AcH 505 at SF (Additional file 6). Among the co-inoculation treatment specific DECs, i.e. contigs which were not DE by sole AcH 505 treatment, we identified contigs homologous to A. thaliana LRR-RLP gene AtRLP9, LRR-RLK gene AtCLV1 and TIR-NB-LRR domain protein gene AtTAO1 (Additional file 6).
Discussion
The RNA-Seq approach has been previously used to explore both global transcriptomic responses of oaks to microbes (mutualists and pathogens) [26, 30, 35]. In the study presented here, we applied RNA-Seq analysis to investigate the impact of the MHB AcH 505 on the oak during two rhythmic growth stages and used combined treatment with the EMF P. croceum to disentangle the direct impact of AcH 505 from the one that is modified in case of co-inoculation. Strong response of the microcutting roots and leaves to the AcH 505 treatment was observed at the root-resting stage. It was manifested by a high number of DECs, and it was in part attenuated by the P. croceum treatment. We began to unravel the gene expression networks linking the microcutting perception of AcH 505 to the plant development, and detected a candidate plant immune response network in the roots following the bacterial inoculation. In this way, our results provide foundation for comparative analyses of interactions between oaks and other microorganisms to elucidate fundamental patterns.
Perception of the microorganisms in oak microcuttings
In accordance with our first hypothesis, we found that defence gene expression in the oak microcuttings were induced by AcH 505, but attenuated by co-inoculation with P. croceum (Fig. 4; Additional file 6). This suggests that the defence suppression by P. croceum dominates over the elicitation of defences by AcH 505. For instance, contigs encoding NDR1-like disease resistance protein, chitinases and hevein were up-regulated genes upon AcH 505 inoculation, but not so upon co-inoculation (Additional file 6), and the GO-term Jasmonic acid mediated signalling pathway was EUC upon AcH 505 inoculation but EUD upon co-inoculation (Fig. 4). The only thus far characterised mechanism how an EMF suppresses plant defences has been presented by Plett, et al. [36]. They showed that Laccaria bicolour produces an effector, mycorrhiza-induced small secreted protein 7 (LbMISSP7) that promotes mycorrhiza formation by blocking plant defence-related JA signalling. P. croceum expresses mycorrhiza-induced small secreted proteins (MISSPs) during EM formation with oak (F. B., S. H., M. T., unpublished), but none of the P. croceum MISSP genes have been functionally characterized. Nevertheless, the suppression by P. croceum observed in co-inoculated plants seems to be a local effect, as it was only detected in roots. Accordingly, Mailänder [20] showed that inoculation of plants with P. croceum did not facilitate powdery mildew infection of oak leaves. In contrast, the enhanced defence gene expression in AcH 505-inoculated plants was also observed in the leaves, suggesting a systemic effect. This supports our previous finding that inoculating pedunculate oak roots with AcH 505 elicits a defence response against M. alphitoides in the leaves [26].
In accordance with our second hypothesis, the gene expression data demonstrated that the transcriptional response of the oak is more active during SF, the period when the roots are not growing. The oak gene expression response to AcH 505 in SF includes DECs encoding resistance proteins, involved in pathogen recognition and subsequent activation of innate immune responses. Candidate LRR containing defence response regulator genes had a particularly SF-specific gene expression (Additional file 6). The strongest induction, 16 to 70-fold expression levels in SF roots and leaves upon AcH 505 and co-treatment, was observed for the LRR-NB-ARC domain protein contig with close homology to AT3G14460 (Table 1). AT3G14460 is a putative disease resistance gene in A. thaliana, and functional characterisation of the oak gene would now prove its role in the recognition of AcH 505.
Detailed analysis of the enriched LRR Pfam terms identified in our experiment in both treatments in roots and leaves during SF (Fig. 3) revealed differential expression of transmembrane RLKs and RLPs (Additional file 6). Some of the LRR-containing receptor transcripts identified in this study were DE in AcH 505- and co-inoculated oaks, suggesting a function as important core genes and central response regulators of the MHB. Among them, a homologue with implicated role in defence against pathogens in A. thaliana [37], an RLK contig homologous to AT3G47110, was strongly (up to 70-fold) up-regulated in roots at SF and at RF by both treatments. The induction at both stages of growth indicates that this candidate RLK may be an important core element of AcH 505 signalling. The strongest up-regulation (up to 80-fold) in leaves occurred for an RLK contig homologous to disease resistance family protein AT2G34930. The AT2G34930 gene is induced in A. thaliana upon infection by the biotrophic oomycete downy mildew pathogen Hyaloperonospora arabidopsidis [37]. These strongly up-regulated transcripts we identified are probably connected to recognition events and maintenance of the interaction with AcH 505.
Transcription factors in plant response to the microorganisms
Crucial part of the plant response to microorganisms and abiotic stressors is mediated by transcriptional control, which is implemented, in large extent, by three families of TFs: ethylene-responsive-element-binding factors (ERF), basic-domain leucine-zipper (bZIP) and WRKY proteins [38]. We presented evidence of DECs among members of these three TF families, occurring in the response of the oak to AcH 505 and/or co-inoculation (Additional file 6). A novel observation for the AcH 505-interaction was the identification of DE ERF contigs amongst the core transcriptome affected by both treatments (Additional file 6). In EM, ET signalling may be involved in control of the depth of colonisation of the plant apoplast by the EMF [39]. Our data indicates that as a MHB, AcH 505 may affect this process by the modification of ERF expression. For instance, ERF8 contig was up-regulated in roots by the interaction with AcH 505 as well as by co-inoculation. Interestingly, the homologue in A. thaliana, AtERF8 (AT1G53170) is a participant in a signalling node of three interacting ERFs, which appear to negatively regulate chitin signalling in defences against fungi, but positively regulate SA signalling in plant defences against the bacterial pathogen Pseudomonas syringae [40]. If the ERF8 homolog of oak shares the functional properties of AtERF8, warrants further analysis.
In tobacco, AP2/ERF subfamily dehydration-responsive element-binding protein gene DREB1B overexpression confers improved abiotic and biotic stress tolerance [41] and in our analysis, the DREB1B expression increased in roots and leaves upon the treatment with AcH 505. In contrast, the gene expression data of Lesur, et al. [42] suggests that candidate DREB1A protein of Q. robur plays a role in the maintenance of bud dormancy, and it thus seems that the DREB subfamily of TFs deserves further attention in oaks.
In A. thaliana, a subgroup of bZIPs is involved in an abscisic acid (ABA) dependent signal transduction pathway under drought and high-salinity conditions, but also in biotic interactions [38]. The induction of oak bZIPs by AcH 505 treatment (Additional file 6) indicates a stress or defence response upon AcH 505 inoculation. The induced bZIPs encoded homologs of central ABA signalling associated TFs, in particular the predicted bZIP63 and GBF3 proteins. From these bZIPs, bZIP63 action relates sugar signalling to ABA synthesis and ABA sensitivity [43], whereas GBF3 is induced by ABA and plays a role in plant response to drought stress and ABA [44] in A. thaliana. We have previously observed that ABA defence pathway may play a role in AcH 505-based priming of host responses against powdery mildew infection [26], and the role of ABA mediated gene expression changes is supported by our data on light-harvesting chlorophyll a/b-binding protein (LHCB) expression as well. The LHCBs are apoproteins of the light-harvesting complexes of chloroplasts, and they are expressed ubiquitously in different plant tissues including roots [45]. Expression of the LHCB genes is modulated by various environmental and developmental cues, including the inducer light [46], and the repressors oxidative stress and ABA [47, 48]. The down-regulation of LHCB expression in roots can be interpreted as a stimulation of normal, darkness related down-regulation, but in leaves, the decrease in LHCB transcript levels suggests increased levels of stress, particularly upon AcH 505 inoculation. The current data further indicates that the suggested induction of stress by AcH 505 is attenuated by P. croceum, since none of the contigs associated with the bZIP category were induced, and only two out of of eleven AcH 505-suppressed LHCB contigs were suppressed upon co-inoculation in leaves at SF (Additional file 6). This raises the question as to how AcH 505, powdery mildew and P. croceum treatments would affect the response of oak to abiotic stress and affect ABA signalling.
Our third TF group of interest, the large family of WRKY proteins plays key roles mediating the regulation of plant defence and abiotic stress transcription and controlling gene expression at leaf senescence [38]. Among the AcH 505-induced WRKYs, homologs of three WRKYs which play roles in signal integration in A. thaliana were detected. WRKY70 protein activates SA-induced genes but represses JA-responsive genes, i.e. controls how the signals converge from these mutually antagonistic pathways [49], and WRKY50 proteins mediate SA-mediated repression of JA signalling [50]. The convergence of signalling pathways is also mediated by WRKY57 protein, which in A. thaliana is a repressor in JA-induced leaf senescence, and a common component of the JA- and auxin-mediated signalling pathways [51]. These results underline the urgency to characterise SA and hormonal signalling pathways in the oak system.
Differential representation of transcripts related to growth and differentiation
The DECs identified suggest that AcH 505 treatment may support the processes of growth and differentiation in oak. Deeper analysis of the DE members of oak AP2/ERF subfamily (Additional file 6) identified a candidate SHINE clade of AP2 domain TF contig. The contig was induced by both treatments in roots and leaves. The corresponding gene AtSHN2 in A. thaliana regulates wax biosynthesis, pectin metabolism and cell wall structure [52], and the induction of a AtSHN2 homolog by both treatments in oak microcuttings might thus be related to the regulation of plant development. Changes in plant growth rates and patterns induced by bacteria and mycorrhizal fungi may be due to associated modifications of the main signalling pathways mediated by not only ET, but also auxin [53]. A homologue of BAK1-interacting RLK1 gene (BIR1, AT5G48380), which stimulates the expression of auxin response regulators in A. thaliana [54] was induced by AcH 505 and a homologue of A. thaliana AIR9 (AT2G34680) gene, an auxin-induced microtubule associated protein which is part of the mechanism positioning the direction of cell division [55], was induced by both AcH 505 and co-inoculation (Additional file 6).
Exordium (EXO) and Exordium-like genes encode cell wall proteins [56], and are structurally related to the tobacco Phosphate-induced protein 1 gene, which is re-activated in cultured cells following release from cell cycle arrest caused by phosphate starvation [57]. Enriched Pfam term PIP1 was associated with increased transcription levels of candidate EXO contigs in both AcH 505 and co-inoculation. In A. thaliana, the EXO proteins connect the extracellular carbon status to growth [58], regulate cell division activity in roots [59], and are essential for cell expansion in leaves [60]. In oak roots, the expression of EXO is upregulated at RF by AcH 505 and co-inoculation, suggesting that the treatments may affect cell division activity of oak roots.
The Pfam term Xyloglucan endo-transglycosylase (XET) was enriched for up-regulated contigs in RF in both treatments (Fig. 3), and for instance, one of the oak XET transcripts was induced by AcH 505 alone and together with P. croceum in RF roots (Table 1). In contrast, the Pfam term XET was enriched for down-regulated contigs in SF in both treatments (Fig. 3), and for instance, the oak XET transcripts with homology to the AtXTH23 gene were down-regulated by both treatments in SF roots (Additional file 6). XET transcripts [61] and activities [62] are largely localized in the root elongation zone, and XET activity is required for the maintenance of root elongation [63]. Thus, since the RF developmental stage coincides with maximal root elongation rate [18], and SF with growth cessation, this association indicates that AcH 505 may enhance root elongation process in oaks during RF and suppress root elongation in SF.
Reversed pattern was observed for the Pfam term Beta-galactosidase, enriched for up-regulated contigs in SF in roots and RF in leaves, and enriched for down-regulated contigs in RF in roots and SF in leaves. The homology of the predicted oak proteins to plant beta galactosidases indicates that they may support tissue differentiation by the modification of cell wall galactans. Of the homologs, A. thaliana BGAL12 (AT4G26140) is localised within cell walls [64], and associated with root differentiation [65]. This association suggests that AcH 505 might stimulate the maturation of cell walls in the rest stages. This is also supported by the induction of an oak peroxidase transcript by AcH 505 alone and together with P. croceum (Table 1). The oak peroxidase contig is orthologous to the product of A. thaliana gene AtPER10 (AT1G49570), known to be up-regulated after ET treatment [66]. Increases in ET and peroxidase activities are both probably involved in the stimulation of peroxidase-mediated crosslinking in the cell wall, which prevents cell expansion [66]. Thus, during SF the up-regulation of peroxidases in oak roots may also contribute to cell wall maturation.
Together, these results suggest an interactive relationship between oak cell wall dynamics and plant-AcH 505 interactions. A similar connection has been suggested for mycorrhizal symbioses in oaks [30, 35] and Populus [39]. Detailed light and electron microscopical analyses of the suppression of Norway spruce root infection by the fungal pathogen Heterobasidion abietinum by the plant-protecting bacterium Streptomyces strain GB 4–2 [67] revealed how the cell walls were reinforced in the lateral roots of co-inoculated plants, preventing the colonisation of the spruce vascular cylinder by the fungal pathogen. In contrast, upon Streptomyces AcH 505-Heterobasidion abietinum-co-inoculation of Norway spruce, the lateral root cell wall structures were not altered [68], and root anatomical analyses were therefore not included in this work. But the current gene expression data indicates that in the oak system, cell biological analysis of the AcH 505-P croceum-interaction is also warranted.
Conclusions
Rhythmic growth affected the gene expression response of oak to the interacting organisms, with much weaker molecular response in RF in terms of the number of DECs. This might indicate that the physiology in roots is rather devoted to growth at RF while the processes directed towards interactions are in most part attenuated. Future work will show if the repressed gene expression response consequently interferes with elicitation of defence responses or symbiosis related signalling in RF. Only few contigs were DE by both AcH 505 and co-inoculation, indicating that oak coordinates its gene expression responses to AcH 505 in the presence and absence of the EMF by induction of a few, specific MAMP receptors and TFs, i.e. candidate “core genes” of AcH 505 response. Our gene expression data further indicate a pivotal role of the cell wall as the responsive element in responses to AcH 505 alone and during co-inoculation with the EMF. Future work needs to focus on the identified candidate genes to verify their involvement in these processes. Clearly, there is also an urgent need to associate these oak genes with hormonal signalling pathways, interactions with insects and pathogens, and with the impact of abiotic environment.
Methods
The soil-based culture system
The pedunculate oak clone DF159 (Quercus robur L.) was micropropagated and rooted according to Herrmann, et al. [69], then cultivated in soil-based microcosms by placing rooted microcuttings in Petri dishes filled with gamma-sterilized soil as previously described [30]. The conditions for bacterial and fungal cultivation and plant inoculation were as described by Tarkka, et al. [30]. Briefly, Piloderma croceum J. Erikss. & Hjortst. Strain 729 (DSM- 4824) inoculum was produced by inoculating a substrate mixture of vermiculite and sphagnum peat with a 2-week-old liquid fungal culture. It was inoculated at establishment of the microcosm by mixing it 1:1 (v/v) with the gamma-sterilized soil. Streptomyces sp. AcH 505 was originally isolated from the soil around Norway spruce mycorrhizas in Haigerloch, Germany [70] and maintained on ISP2 agar medium [71]. For the experiment, the culture system was inoculated with 2.5 × 107 AcH 505 spores at 3 and 7 weeks after establishment of the microcosm. With this form of inoculation, AcH 505 stimulates oak-P. croceum mycorrhiza formation [25], and elicits plant defences in leaves against the powdery mildew Microsphaera alphitoides [26]. After 4 weeks, 5 ml of soil filtrate was added to each culture system to establish a natural microbial community and thereby making the system closer to natural conditions. Soil filtrates were prepared as described by Rosenberg, et al. [72]. Briefly, the microbial filtrate was obtained by suspending the same soil from an oak forest stand that was used as a soil substrate for the microcosms in water and filtering it through 5.0 and 1.2 μm paper filters, thereby excluding Protozoa.
The oak microcuttings were cultivated for 8 weeks in climate chambers at 23 °C with a 16/8 h day/night (photosynthetic photon flux density of 180 μmol m−2 s−1 ), before the tissues were harvested. After harvest, tissues were immediately submerged in liquid nitrogen and stored at −80 °C. Harvest times were based on published effects of AcH 505 on EM formation and plant growth [25, 26].
For our investigations we only used plants that were in the developmental root flush (RF) and shoot flush (SF) stages, corresponding respectively to bud swelling stage B and leaf expansion stage D according to Herrmann, et al. [18]. The experimental design resulted in six treatments: 3 (no inoculation/AcH 505/AcH 505 + P. croceum) × 2 (RF/SF).
RNA extraction and transcript quantitation by Illumina sequencing
For transcriptomic analyses, lateral roots and leaves were used. Source and sink leaves were used for plants during RF and SF respectively. Samples from 2–3 plants subjected to each treatment were pooled and homogenized under liquid nitrogen. Total RNA was isolated from 36 pools (3 treatments × 2 tissues × 2 developmental stages × 3 replicates) using the MasterPure Plant RNA Purification Kit (Epicentre, Hessisch Oldendorf, Germany) with 100 mg of root and 50 mg of leaf material per extraction. RNA quality and quantity were verified using gel electrophoresis, a NanoDrop 1000 spectrophotometer and an Agilent 2100 Bioanalyzer prior to Illumina sequencing analysis at the Beijing Genomics Institute (Hong Kong, China). 100 bp paired-end Illumina Truseq version 2 libraries were constructed and sequenced using the Illumina HiSeq2000 sequencing platform, and the sequence data was deposited as fastq files to the NCBI Short Read Archive, linked to a report specific BioProject termed PRJNA280092.
Read processing and analysis of differential expression
Reads were processed following Tarkka, et al. [30]. Briefly, low quality sequences and sequencing artefacts were removed with SeqClean (http://sourceforge.net/projects/seqclean/files/) and low quality sequencing ends were trimmed with a custom Java script. Short sequences (<50 bp) and sequences lacking paired-end information were discarded. The processed Illumina reads were aligned against the reference transcriptome OakContigDF159.1 [30] by Bowtie [73] and quantified by RSEM [74]. Fold-changes in gene expression were calculated by pairwise comparisons using the edgeR function [75] implemented in the Bioconductor package [76]. In these comparisons, negative binomial models are fitted to the transcript abundancies determined by RSEM. Contigs were considered differentially expressed (DE) when the Benjamini-Hochberg adjusted P-value of this fit was less than 1 %. Blast2GO was used to get a description for each contig based on up to 20 hits against NCBI NR database in a blastx search (E-value 1e-5). Protein sequences from Arabidopsis thaliana TAIR database were downloaded to perform a blastx search of DF159.1 and to assign homologue proteins from A. thaliana to each contig. Only hits with an E-value of at least 1e-5 were taken into account. The best A. thaliana protein hit for each oak contig was determined by taking the A. thaliana protein exhibiting the largest percent identity to the contig in the local alignment.
Gene Ontology (GO) [77] enrichment analysis was performed with the Bioconductor package GOseq [78]. GOseq performs a statistical test based on a hypergeometric distribution to determine if in a given list of DE tags (e.g. genes or contigs) tags assigned to a certain category (e.g. GO terms) are significantly enriched, i.e. if they occur more frequently than expected by chance. Thereby GOseq adjusts the estimation of the P-value for tag-length. We used the capability of GOseq to perform enrichment analyses for a second type of categories, Protein families (Pfam) [79]. The OakContigDF159.1 reference library, GO annotations as well as best blast hits of each contig have been deposited at www.trophinoak.de.
Real-time-quantitative reverse transcriptase-PCR (qRT-PCR) primer design and reactions
Differential gene expression data obtained from the Illumina analyses were validated by qRT-PCR analysis of the expression of 10 genes in leaf samples from control and co-inoculated plants. Primer pairs were constructed using the OakContigDF159.1 assembly as a reference and tested for functionality, amplicon size, specificity and efficiency as previously described [30]. Sequences of constructed primer pairs are listed in Additional file 7. The qRT-PCR reactions were performed as described by Tarkka, et al. [30]. Briefly, using an iScript One-Step RT-PCR Kit with SYBR Green (Bio-Rad) and 18S rRNA as the reference gene, transcript abundances in the leaf samples were determined based on their Ct values using the Relative Expression Software Tool [REST, 80]. The coefficient of variation was used as a reproducibility indicator, with a maximal value of 6.0. Differential gene expression was determined by a randomisation test implemented in REST.
Availability of supporting data
Supporting data are included as Additional files.
The OakContigDF159.1 reference library, GO annotations as well as best blast hits of each contig have been deposited at www.trophinoak.de.
The sequence data as original fastq files is deposited at NCBI Short Read Archive, linked to a report specific BioProject termed PRJNA280092 (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA280092).
Acknowledgements
We thank Kerstin Hommel for help with the bacterial and fungal cultures, Ines Krieg and Barbara Krause for oak micropropagation and the participants of the TrophinOak project (www.trophinoak.de) for excellent collaboration and useful discussions. Support from the grants TA 290/4-1 and BU 941/20-1 provided by the German Science Foundation (DFG) is gratefully acknowledged, as well as a Flexible Pool Grant (number 50170649_#3) of the German Centre for Integrative Biodiversity Research (iDiv) Halle – Jena – Leipzig. FK was kindly supported by the Helmholtz Impulse and Networking Fund through the Helmholtz Interdisciplinary Graduate School for Environmental Research (HIGRADE).
Abbreviations
- ABA
Abscisic acid
- bZIP
Basic-domain leucine-zipper
- CHLBP
Chlorophyll A-B binding protein
- DE
Differentially expressed
- DEC
Differentially expressed contig
- EDC
Enriched in down-regulated contigs
- EM
Ectomycorrhiza
- EMF
Ectomycorrhizal fungus
- ERF
Ethylene-responsive element binding factor
- ET
Ethylene
- EUC
Enriched in up-regulated contigs
- FDA
Fasciclin domain arabinogalactan protein
- GDSL
GDSL-like Lipase/Acylhydrolase
- GO
Gene Ontology
- JA
Jasmonic acid
- LHCB
Light-harvesting chlorophyll a/b-binding protein
- LRR
Leucine rich repeat
- LRR-NB
Leucine rich repeat-nuleotide binding
- LTP
Probable lipid transfer protein
- MHB
Mycorrhiza helper bacterium
- Pfam
Protein family
- PIP1
Phosphate-induced protein 1
- qRT-PCR
Real-time-quantitative reverse transcriptase-PCR
- RF
Root flush
- RLK
Receptor-like kinase
- RLP
Receptor-like protein
- SF
Shoot flush
- TF
Transcription factor
- SA
Salicylic acid
Additional files
Results of Illumina sequencing of Quercus robur DF159 cDNA libraries. (XLS 50 kb)
Mapping of reads on the OakContigDF159.1 reference transcriptome. (DOCX 72 kb)
Results of qRT-PCR analysis. (DOCX 16 kb)
Differential gene expression levels in numbers. (DOCX 18 kb)
The most significantly enriched Pfam and GO terms. (XLSX 31 kb)
DECs associated with enriched Pfam and GO terms. (XLSX 57 kb)
Quantitative polymerase chain reaction primer sequences. (DOCX 15 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MT, FK, SH designed the research; FK performed the research; FK, LF, MB, MT analysed the data; FK, MT wrote the paper. FB is the principal investigator of the TrophinOak project. All authors read and approved the final manuscript.
Contributor Information
Florence Kurth, Email: florence.kurth@ufz.de.
Lasse Feldhahn, Email: lasse.feldhahn@ufz.de.
Markus Bönn, Email: markus.boenn@ufz.de.
Sylvie Herrmann, Email: sylvie.herrmann@ufz.de.
François Buscot, Email: francois.buscot@ufz.de.
Mika T. Tarkka, Email: mika.tarkka@ufz.de
References
- 1.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37(5):634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 2.Smith SE, Read DJ. Mycorrhizal symbiosis. London: Academic; 2008. [Google Scholar]
- 3.Frey-Klett P, Garbaye J, Tarkka M. The mycorrhiza helper bacteria revisited. New Phytol. 2007;176(1):22–36. doi: 10.1111/j.1469-8137.2007.02191.x. [DOI] [PubMed] [Google Scholar]
- 4.Riedlinger J, Schrey SD, Tarkka MT, Hampp R, Kapur M, Fiedler HP. Auxofuran, a novel metabolite that stimulates the growth of fly agaric, is produced by the mycorrhiza helper bacterium Streptomyces strain AcH 505. Appl Environ Microbiol. 2006;72(5):3550–3557. doi: 10.1128/AEM.72.5.3550-3557.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brulé C, Frey-Klett P, Pierrat JC, Courrier S, Gerard F, Lemoine MC, Rousselet JL, Sommer G, Garbaye J. Survival in the soil of the ectomycorrhizal fungus Laccaria bicolor and the effects of a mycorrhiza helper Pseudomonas fluorescens. Soil Biol Biochem. 2001;33(12–13):1683–1694. doi: 10.1016/S0038-0717(01)00090-6. [DOI] [Google Scholar]
- 6.Vivas A, Barea JM, Azcón R. Brevibacillus brevis isolated from cadmium- or zinc-contaminated soils improves in vitro spore germination and growth of Glomus mosseae under high Cd or Zn concentrations. Microb Ecol. 2005;49(3):416–424. doi: 10.1007/s00248-004-0044-4. [DOI] [PubMed] [Google Scholar]
- 7.Duponnois R. Les bacteries auxilaires de la mycorrhization du Douglas (Pseudotsuga menziessii (Mirb.) Franco) par Laccaria laccata souche S238. France: University of Nancy 1; 1992. [Google Scholar]
- 8.Toro M, Azcon R, Barea J. Improvement of arbuscular mycorrhiza development by inoculation of soil with phosphate-solubilizing rhizobacteria to improve rock phosphate bioavailability ((sup32) P) and nutrient cycling. Appl Environ Microbiol. 1997;63(11):4408–4412. doi: 10.1128/aem.63.11.4408-4412.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frey-Klett P, Garbaye J. Mycorrhiza helper bacteria: a promising model for the genomic analysis of fungal-bacterial interactions. New Phytol. 2005;168(1):4–8. doi: 10.1111/j.1469-8137.2005.01553.x. [DOI] [PubMed] [Google Scholar]
- 10.Buscot F, Herrmann S: At the frontier between Basidiomycotes and plants: reciprocal interactions between mycorrhiza formation and root development in an in vitro system with oaks and Hymenomycetes. In: Frontiers in Basidiomycote mycology. Edited by AgererR, Piepenbring M, Blanz P. Eching, Germany: IHW-Verlag & Verlagsbuchhandlung; 2004:361–376.
- 11.Lavarenne S. Croissance rythmique de quelques espèces de chênes cultivés en chambres climatisées. Comptes Rendus Hebdomadaires Des Seances De L Academie Des Sciences Serie D. 1966;262(3):358. [Google Scholar]
- 12.Champagnat P, Payan E, Champagnat M, Barnola P, Lavarenne S, Bertholon C. La croissance rythmique de jeunes chenes peduncolees cultives en conditions controlees et uniformes. Naturalia monspeliensia - Colloque international sur l'arbre. 1986;303–337.
- 13.Alatou D, Barnola P, Lavarenne S, Gendraud M. Caractérisation de la croissance rythmique du chêne pédonculé. Plant Physiol Biochem. 1989;27(2):275–280. [Google Scholar]
- 14.Alaoui Sossé B, Parmentier C, Dizengremel P, Barnola P. Rhythmic growth and carbon allocation in Quercus robur. 1. Starch and sucrose. Plant Physiol Biochem. 1994;32(3):331–339. [Google Scholar]
- 15.Le Hir R, Leduc N, Jeannette E, Viemont J-D, Pelleschi-Travier S. Variations in sucrose and ABA concentrations are concomitant with heteroblastic leaf shape changes in a rhythmically growing species (Quercus robur) Tree Physiol. 2006;26(2):229–238. doi: 10.1093/treephys/26.2.229. [DOI] [PubMed] [Google Scholar]
- 16.Willaume M, Pagès L. How periodic growth pattern and source/sink relations affect root growth in oak tree seedlings. J Exp Bot. 2006;57(4):815–826. doi: 10.1093/jxb/erj059. [DOI] [PubMed] [Google Scholar]
- 17.Costes E, García-Villanueva E, Jourdan C, Regnard J, Guédon Y. Co-ordinated growth between aerial and root systems in young apple plants issued from in vitro culture. Ann Bot. 2006;97(1):85–96. doi: 10.1093/aob/mcj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann S, Munch JC, Buscot F. A gnotobiotic culture system with oak microcuttings to study specific effects of mycobionts on plant morphology before, and in the early phase of, ectomycorrhiza formation by Paxillus involutus and Piloderma croceum. New Phytol. 1998;138(2):203–212. doi: 10.1046/j.1469-8137.1998.00105.x. [DOI] [PubMed] [Google Scholar]
- 19.Angay O, Fleischmann F, Recht S, Herrmann S, Matyssek R, Oßwald W, Buscot F, Grams TE. Sweets for the foe–effects of nonstructural carbohydrates on the susceptibility of Quercus robur against Phytophthora quercina. New Phytol. 2014;203(4):1282–1290. doi: 10.1111/nph.12876. [DOI] [PubMed] [Google Scholar]
- 20.Mailänder S. Einfluss des Blattpathogens Microsphaera alphitoides, des Ektomykorrhizapilzes Piloderma croceum und des Streptomyceten AcH 505 auf Transkriptom, Physiologie und Morphologie der klonalen Eichenstecklinge Q. robur DF159. Tübingen, Germany: University of Tübingen; 2014. [Google Scholar]
- 21.Faeth SH, Hammon KE. Fungal endophyres in oak trees: experimental analyses of interactions with leafminers. Ecology. 1997;78(3):820–827. doi: 10.1890/0012-9658(1997)078[0820:FEIOTE]2.0.CO;2. [DOI] [Google Scholar]
- 22.Tack AJ, Gripenberg S, Roslin T. Cross‐kingdom interactions matter: fungal-mediated interactions structure an insect community on oak. Ecol Lett. 2012;15(3):177–185. doi: 10.1111/j.1461-0248.2011.01724.x. [DOI] [PubMed] [Google Scholar]
- 23.Leski T, Pietras M, Rudawska M. Ectomycorrhizal fungal communities of pedunculate and sessile oak seedlings from bare-root forest nurseries. Mycorrhiza. 2010;20(3):179–190. doi: 10.1007/s00572-009-0278-6. [DOI] [PubMed] [Google Scholar]
- 24.Garbaye J, Churin JL, Duponnois R. Effects of substrate sterilization, fungicide treatment, and mycorrhization helper bacteria on ectomycorrhizal formation of pedunculate oak (Quercus robur) inoculated with Laccaria laccata in 2 peat bare root nurseries. Biol Fertil Soils. 1992;13(1):55–57. doi: 10.1007/BF00337239. [DOI] [Google Scholar]
- 25.Kurth F, Zeitler K, Feldhahn L, Neu TR, Weber T, Krištůfek V, Wubet T, Herrmann S, Buscot F, Tarkka MT. Detection and quantification of a mycorrhization helper bacterium and a mycorrhizal fungus in plant-soil microcosms at different levels of complexity. BMC Microbiol. 2013;13:205. doi: 10.1186/1471-2180-13-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurth F, Mailänder S, Bönn M, Feldhahn L, Herrmann S, Große I, Buscot F, Schrey SD, Tarkka MT. Streptomyces-induced resistance against oak powdery mildew involves host plant responses in defense, photosynthesis, and secondary metabolism pathways. Mol Plant-Microbe Interact. 2014;27(9):891–900. doi: 10.1094/MPMI-10-13-0296-R. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann S, Buscot F. Cross talks at the morphogenetic, physiological and gene regulation levels between the mycobiont Piloderma croceum and oak microcuttings (Quercus robur) during formation of ectomycorrhizas. Phytochemistry. 2007;68(1):52–67. doi: 10.1016/j.phytochem.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 28.Krüger A, Frettinger P, Herrmann S, Buscot F, Oelmüller R. Identification of premycorrhiza‐related plant genes in the association between Quercus robur and Piloderma croceum. New Phytol. 2004;163(1):149–157. doi: 10.1111/j.1469-8137.2004.01091.x. [DOI] [PubMed] [Google Scholar]
- 29.Frettinger P, Derory J, Herrmann S, Plomion C, Lapeyrie F, Oelmüller R, Martin F, Buscot F. Transcriptional changes in two types of pre-mycorrhizal roots and in ectomycorrhizas of oak microcuttings inoculated with Piloderma croceum. Planta. 2007;225(2):331–340. doi: 10.1007/s00425-006-0355-4. [DOI] [PubMed] [Google Scholar]
- 30.Tarkka MT, Herrmann S, Wubet T, Feldhahn L, Recht S, Kurth F, Mailänder S, Bönn M, Neef M, Angay O. OakContigDF159. 1, a reference library for studying differential gene expression in Quercus robur during controlled biotic interactions: use for quantitative transcriptomic profiling of oak roots in ectomycorrhizal symbiosis. New Phytol. 2013;199(2):529–540. doi: 10.1111/nph.12317. [DOI] [PubMed] [Google Scholar]
- 31.Newman M-A, Sundelin T, Nielsen JT, Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Frontiers Plant Sci. 2013;4:139. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukasik E, Takken FL. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol. 2009;12(4):427–436. doi: 10.1016/j.pbi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411(6839):826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 34.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2012;1819(2):86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Sebastiana M, Vieira B, Lino-Neto T, Monteiro F, Figueiredo A, Sousa L, Pais MS, Tavares R, Paulo OS. Oak root response to ectomycorrhizal symbiosis establishment: RNA-Seq derived transcript identification and expression profiling. Plos One. 2014;9(5) doi: 10.1371/journal.pone.0098376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plett JM, Daguerre Y, Wittulsky S, Vayssières A, Deveau A, Melton SJ, Kohler A, Morrell-Falvey JL, Brun A, Veneault-Fourrey C. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc Natl Acad Sci. 2014;111(22):8299–8304. doi: 10.1073/pnas.1322671111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hok S, Danchin EG, Allasia V, Panabieres F, Attard A, Keller H. An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 2011;34(11):1944–1957. doi: 10.1111/j.1365-3040.2011.02390.x. [DOI] [PubMed] [Google Scholar]
- 38.Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5(5):430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 39.Plett JM, Khachane A, Ouassou M, Sundberg B, Kohler A, Martin F. Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between Laccaria bicolor and Populus roots. New Phytol. 2014;202(1):270–286. doi: 10.1111/nph.12655. [DOI] [PubMed] [Google Scholar]
- 40.Son GH, Wan J, Kim HJ, Nguyen XC, Chung WS, Hong JC, Stacey G. Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol Plant-Microbe Interact. 2012;25(1):48–60. doi: 10.1094/MPMI-06-11-0165. [DOI] [PubMed] [Google Scholar]
- 41.Gutha LR, Reddy AR. Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol Biol. 2008;68(6):533–555. doi: 10.1007/s11103-008-9391-8. [DOI] [PubMed] [Google Scholar]
- 42.Lesur I, Le Provost G, Bento P, Da Silva C, Leplé J-C, Murat F, Ueno S, Bartholomé J, Lalanne C, Ehrenmann F, et al. The oak gene expression atlas: insights into Fagaceae genome evolution and the discovery of genes regulated during bud dormancy release. BMC Genomics. 2015;16(1):112. doi: 10.1186/s12864-015-1331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matiolli CC, Tomaz JP, Duarte GT, Prado FM, Del Bem LEV, Silveira AB, Gauer L, Corrêa LGG, Drumond RD, Viana AJC. The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiol. 2011;157(2):692–705. doi: 10.1104/pp.111.181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu G, Paul A-L, McCarty DR, Ferl RJ. Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell Online. 1996;8(5):847–857. doi: 10.1105/tpc.8.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jansson S. The light-harvesting chlorophyll ab-binding proteins. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1994;1184(1):1–19. doi: 10.1016/0005-2728(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 46.Silverthorne J, Tobin EM. Demonstration of transcriptional regulation of specific genes by phytochrome action. Proc Natl Acad Sci. 1984;81(4):1112–1116. doi: 10.1073/pnas.81.4.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staneloni RJ, Rodriguez-Batiller MJ, Casal JJ. Abscisic acid, high-light, and oxidative stress down-regulate a photosynthetic gene via a promoter motif not involved in phytochrome-mediated transcriptional regulation. Mol Plant. 2008;1(1):75–83. doi: 10.1093/mp/ssm007. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y-H, Liu R, Yan L, Liu Z-Q, Jiang S-C, Shen Y-Y, Wang X-F, Zhang D-P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J Exp Bot. 2012;63(3):1095–1106. doi: 10.1093/jxb/err315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell Online. 2004;16(2):319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Q-M, Venugopal S, Navarre D, Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011;155(1):464–476. doi: 10.1104/pp.110.166876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Y, Liang G, Yang S, Yu D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid–and auxin-mediated signaling in jasmonic acid–induced leaf senescence. Plant Cell Online. 2014;26(1):230–245. doi: 10.1105/tpc.113.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 2011;7(5) doi: 10.1371/journal.pgen.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vacheron J, Desbrosses G, Bouffaud M-L, Touraine B, Moënne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dyé F, Prigent-Combaret C. Plant growth-promoting rhizobacteria and root system functioning. Frontiers Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim MH, Kim Y, Kim JW, Lee H-S, Lee WS, Kim S-K, Wang Z-Y, Kim S-H. Identification of Arabidopsis BAK1-associating receptor-like kinase 1 (BARK1) and characterization of its gene expression and brassinosteroid-regulated root phenotypes. Plant Cell Physiol. 2013;54(10):1620–1634. doi: 10.1093/pcp/pct106. [DOI] [PubMed] [Google Scholar]
- 55.Buschmann H, Chan J, Sanchez-Pulido L, Andrade-Navarro MA, Doonan JH, Lloyd CW. Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr Biol. 2006;16(19):1938–1943. doi: 10.1016/j.cub.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 56.Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics. 2006;6(1):301–311. doi: 10.1002/pmic.200500046. [DOI] [PubMed] [Google Scholar]
- 57.Sano T, Kuraya Y, Amino S-i, Nagata T. Phosphate as a limiting factor for the cell division of tobacco BY-2 cells. Plant Cell Physiol. 1999;40(1):1–16. doi: 10.1093/oxfordjournals.pcp.a029464. [DOI] [PubMed] [Google Scholar]
- 58.Lisso J, Schröder F, Müssig C. EXO modifies sucrose and trehalose responses and connects the extracellular carbon status to growth. Frontiers Plant Sci. 2013;4:219. doi: 10.3389/fpls.2013.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farrar K, Evans IM, Topping JF, Souter MA, Nielsen JE, Lindsey K. EXORDIUM–a gene expressed in proliferating cells and with a role in meristem function, identified by promoter trapping in Arabidopsis. Plant J. 2003;33(1):61–73. doi: 10.1046/j.1365-313X.2003.01608.x. [DOI] [PubMed] [Google Scholar]
- 60.Schröder F, Lisso J, Lange P, Müssig C. The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol. 2009;9(1):20. doi: 10.1186/1471-2229-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu J, Chen S, Alvarez S, Asirvatham VS, Schachtman DP, Wu Y, Sharp RE. Cell wall proteome in the maize primary root elongation zone. I. Extraction and identification of water-soluble and lightly ionically bound proteins. Plant Physiol. 2006;140(1):311–325. doi: 10.1104/pp.105.070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pritchard J, Hetherington PR, Fry SC, Tomos AD. Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. J Exp Bot. 1993;44(8):1281–1289. doi: 10.1093/jxb/44.8.1281. [DOI] [Google Scholar]
- 63.Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC. Root growth maintenance at low water potentials (increased activity of xyloglucan endotransglycosylase and its possible regulation by abscisic acid) Plant Physiol. 1994;106(2):607–615. doi: 10.1104/pp.106.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gantulga D, Ahn YO, Zhou C, Battogtokh D, Bevan DR, Winkel BSJ, Esen A. Comparative characterization of the Arabidopsis subfamily a1 β-galactosidases. Phytochemistry. 2009;70(17–18):1999–2009. doi: 10.1016/j.phytochem.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 65.Albornos L, Martín I, Pérez P, Marcos R, Dopico B, Labrador E. Promoter activities of genes encoding β-galactosidases from Arabidopsis a1 subfamily. Plant Physiol Biochem. 2012;60:223–232. doi: 10.1016/j.plaphy.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 66.Markakis MN, De Cnodder T, Lewandowski M, Simon D, Boron A, Balcerowicz D, Doubbo T, Taconnat L, Renou J-P, Höfte H. Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol. 2012;12(1):208. doi: 10.1186/1471-2229-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lehr NA, Schrey SD, Hampp R, Tarkka MT. Root inoculation with a forest soil streptomycete leads to locally and systemically increased resistance against phytopathogens in Norway spruce. New Phytol. 2008;177:965–976. doi: 10.1111/j.1469-8137.2007.02322.x. [DOI] [PubMed] [Google Scholar]
- 68.Lehr NA, Schrey SD, Bauer R, Hampp R, Tarkka MT. Suppression of plant defence response by a mycorrhiza helper bacterium. New Phytol. 2007;174(4):892–903. doi: 10.1111/j.1469-8137.2007.02021.x. [DOI] [PubMed] [Google Scholar]
- 69.Herrmann S, Oelmuller R, Buscot F. Manipulation of the onset of ectomycorrhiza formation by indole-3-acetic acid, activated charcoal or relative humidity in the association between oak microcuttings and Piloderma croceum: influence on plant development and photosynthesis. J Plant Physiol. 2004;161(5):509–517. doi: 10.1078/0176-1617-01208. [DOI] [PubMed] [Google Scholar]
- 70.Maier A, Riedlinger J, Fiedler H-P, Hampp R. Actinomycetales bacteria from a spruce stand: characterization and effects on growth of root symbiotic, and plant parasitic soil fungi in dual culture. Mycol Prog. 2004;3:129–136. doi: 10.1007/s11557-006-0083-y. [DOI] [Google Scholar]
- 71.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 72.Rosenberg K, Bertaux J, Krome K, Hartmann A, Scheu S, Bonkowski M. Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J. 2009;3(6):675–684. doi: 10.1038/ismej.2009.11. [DOI] [PubMed] [Google Scholar]
- 73.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young M, Wakefield M, Smyth G, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(D1):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:9. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]


