Table 5.
Identification of candidate diagnostic markers through genotyping sequence-defined markers with whole genome sequencing data from 10 cultivars on genetic linkage map flanking the R gene PhtjR conferring resistance to phomopsis in Lupinus angustifolius
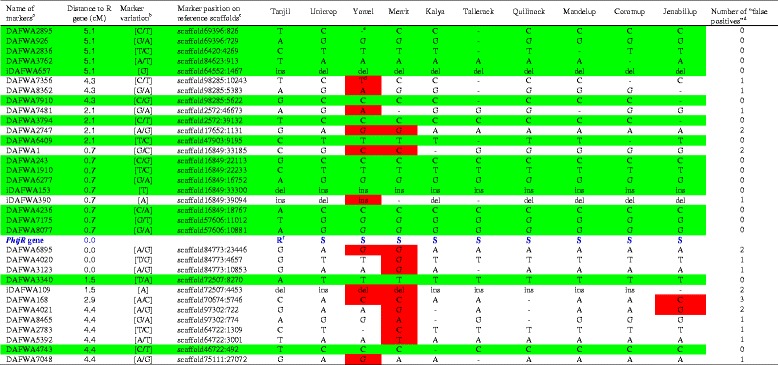
aMarkers showing genotypes completely consistent with PSB disease phenotypes on all 10 cultivars are considered candidate diagnostic markers and are highlighted in green
bTwo nucleotides separated by a stroke line in brackets are SNP markers; nucleotides in brackets without a stroke line are InDel markers
cMarker positions are the nucleotide positions on the reference genome sequence assembly from cultivar Tanjil (Genbank BioProject number PRJNA179231)
dMarkers showing R-allele genotype on cultivars without the R gene Phtj (false positives) are highlighted in red
eMarker sequences missing in genome re-sequencing were recorded as missing data “-”
fGenotypes of R gene PhtjR on sequenced cultivars presented in blue: R = presence of PhtjR gene; S = absence of PhtjR gene [44]
