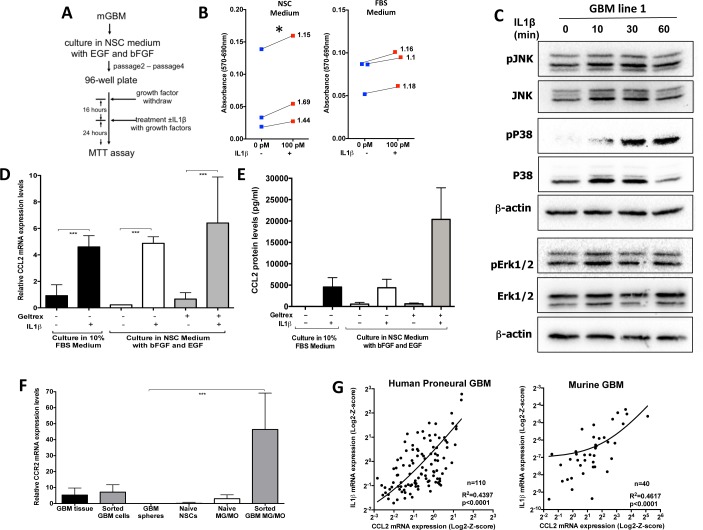
Figure 5. IL1β treatment increases glioma cell growth/viability, activates p38/JNK and NF-κB pathways in GSC-enriching medium, and leads to activation of CCL2 expression.
A) Illustration of the steps for the MTT assay. B) Quantification of the MTT assay performed on GBM spheres, which were cultured in NSC (left) or 10% FBS (right) with or without IL1β 24h later. These data show that while there was no significant increase in the growth in FBS, there was a significant difference in NSC conditions. We used three independent primary tumors to derive three independent glioma lines for two conditions. Blue squares show the growth of the original tumor and they are connected to red squares that represent the growth in response to IL1β; values on the graphs represent fold increase in growth/viability in response to IL1β compare to control growth. An unpaired t-test was used to determine the fold change compare to control, * p < 0.05, 95% CI (1,33; 1,69). C) Immunoblot analysis was performed to examine the p38 MAPK, JNK and ERK1/2 signaling pathways 0, 10, 30 and 60 minutes after 100pM IL1β treatment. The Western blots shown were performed on the GBM 1 and GBM 2 lines (cultured in Geltrex). Similar results were obtained on GBM sphere 3. D) Relative mRNA expression dot plots for CCL2 from glioma cells cultured in different conditions with or without 100pM IL1β treatment, showing that IL1β treatment significantly increased CCL2 mRNA expression, independent of culture conditions. E) The CCL2 protein levels measured by ELISA in the supernatant showed an increase in response to 100pM IL1β treatment. F) Relative mRNA expression dot plots for CCR2 expression from different cell types showing that CCR2 was not expressed in cultured glioma cells when grown in NSC conditions. A one-way ANOVA with Tukey's multiple comparisons test was performed, ***p < 0.001 G) The right graph shows correlation with linear regression from 110 proneural GBM samples for IL1β and CCL2 RNA expression from the TCGA database. A statistically significant positive correlation was found between IL1β and CCL2 RNA expression r=0.63, p<0.0001. Linear regression R2=0.4397, p < 0.001. The left graph shows correlation with linear regression from 40 murine PDGF-B-driven GBM samples for IL1β and CCL2 RNA expression. A statistically significant positive correlation was found between IL1β and CCL2 RNA expression r=0.68, p<0.001. Linear regression R2=0.4617, p < 0.001.