Figure 1. The regulatory network coupling OVOL with miR-200/ZEB.
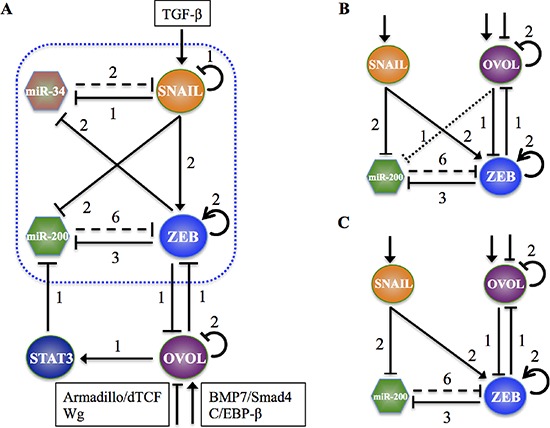
A. The regulatory network coupling OVOL with miR-34/SNAIL and miR-200/ZEB circuits for prostate cancer. The part of the circuit in the dotted box shows the core EMT regulatory network. OVOL forms a mutually inhibitory loop with ZEB, inhibits miR-200 indirectly via STAT3 and is also self-inhibitory. TGF-β activates SNAIL, and BMP7/Smad4 pathway and C/EBP-β activate OVOL, but Wg signaling (Armadillo/dTCF) inhibits OVOL. The coupling of miR-34/SNAIL with miR-200/ZEB does not change the qualitative behavior of miR-200/ZEB circuit [19]. For this reason, in order to simplify the calculations, we didn't consider here the miR-34/SNAIL loop and simply treated SNAIL as an external signal on the miR-200/ZEB/OVOL circuit. B. Effective miR-200/ZEB/OVOL circuit for the case of prostate cancer, where OVOL inhibits both miR-200 and ZEB. C. Effective miR-200/ZEB/OVOL circuit for the case of breast cancer, where OVOL inhibits ZEB but not miR-200. A solid arrow denotes transcriptional activation, and a solid bar denotes transcriptional inhibition. Dashed line indicates microRNA-mediated translational regulation, and dotted line indicates indirect inhibition. The number listed along each line represents the number of binding sites on the promoter region of target genes (See SI section I for details). In B and C, external signals on SNAIL and OVOL denote those shown in A.
