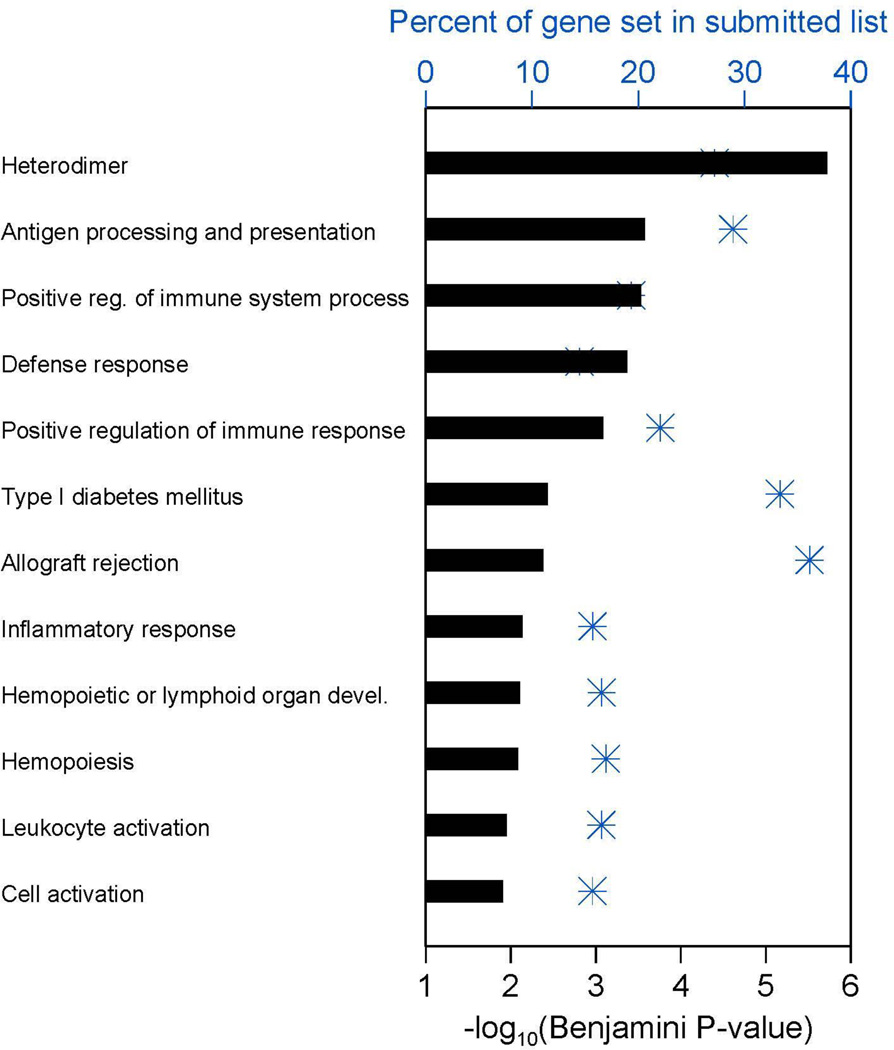
Figure 1. Whole genome microarray reveals functional gene categories that are differentially expressed in PD-L1+ vs. negative melanomas.
In PD-L1+ melanomas, TIL intensity was moderate to severe, while in PD-L1(−) melanomas it was mild. Blue asterisks show the percent of each gene set that occurred in the list submitted to DAVID analysis, using the x-axis scale at the top of the plot. The horizontal bar for each gene set shows –log10 of the corresponding Benjamini-Hochberg multiple comparison adjusted p-value as calculated by DAVID, using the x-axis scale at the bottom of the plot. Functional categories with a gene count ≥10 and a Benjamini p-value ≤0.015 are shown. Additional information, including the specific number of genes in the submitted list present in each DAVID functional category appearing in the plot, is provided in Supplementary Tables S3 and S4.