Figure 2.
DA1, DAR1, and DAR2 Redundantly Regulate Plant Growth and Development in a Context-Dependent Manner.
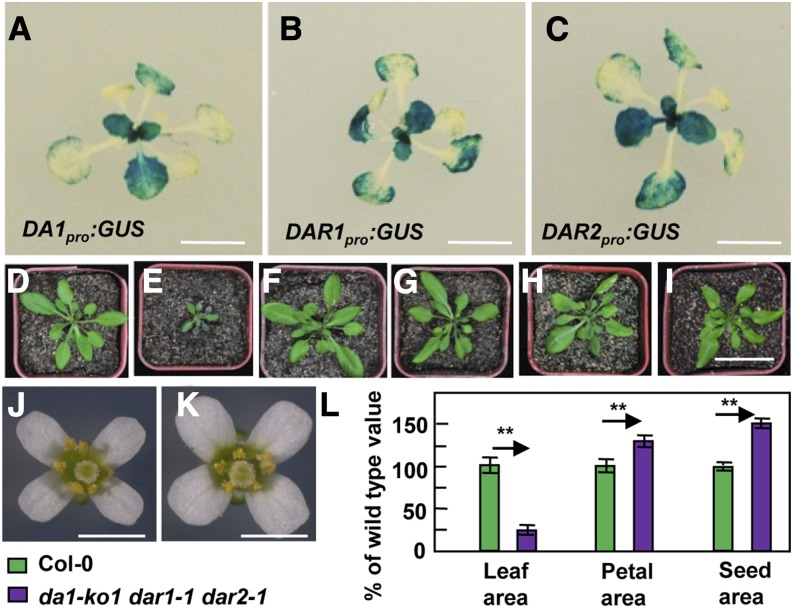
(A) to (C) The expression activity of DA1, DAR1, and DAR2 was monitored by DA1pro:GUS, DAR1pro:GUS, and DAR2pro:GUS transgene expression. Histochemical analysis of GUS activity in 16-d-old plants is shown.
(D) to (I) Thirty-day-old plants of Col-0 (D), da1-ko1 dar1-1 dar2-1 (E), 35S:DA1;da1-ko1 dar1-1 dar2-1 (F), 35S:DAR1;da1-ko1 dar1-1 dar2-1 (G), 35S:DAR2;da1-ko1 dar1-1 dar2-1 (H), and gDAR2;da1-ko1 dar1-1 dar2-1 (I). 35S:DA1;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with the DA1 coding sequence driven by the 35S promoter. 35S:DAR1;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with the DAR1 coding sequence driven by the 35S promoter. 35S:DAR2;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with the DAR2 coding sequence driven by the 35S promoter. gDAR2;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with a DAR2 genomic copy. Expression of DA1, DAR1, DAR2, or gDAR2 rescued the small-plant phenotype of da1-ko1 dar1-1 dar2-1.
(J) and (K) Flowers of Col-0 and da1-ko1 dar1-1 dar2-1 plants.
(L) Leaf area, petal area, and seed area of Col-0 and da1-ko1 dar1-1 dar2-1 plants. Values are given as means ± se relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test).
Bars in (A) to (C) = 2 cm; bar in (D) to (I) = 5 cm; bars in (J) and (K) = 2 mm.