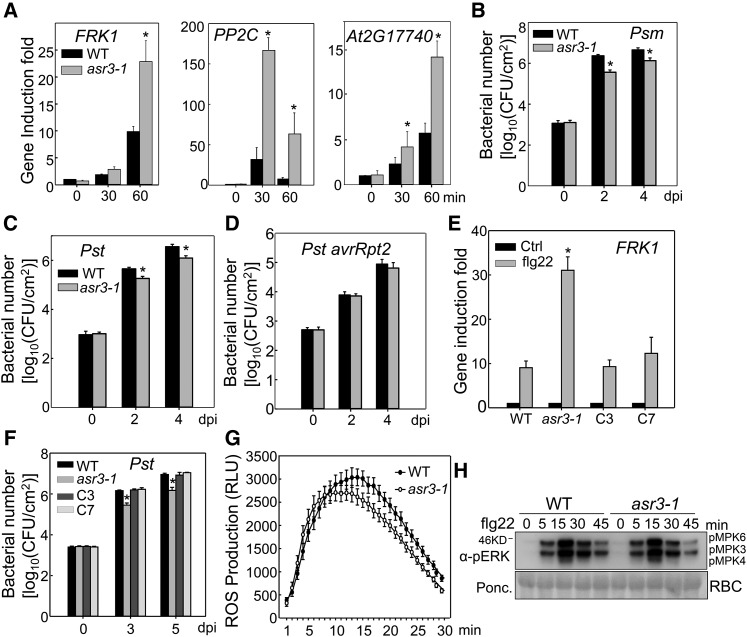
Figure 1.
The asr3 Mutant Displays Enhanced Disease Resistance and Immune Gene Activation.
(A) The flg22-induced marker gene expression in the wild type and asr3-1 mutant. Ten-day-old seedlings were treated with 100 nM flg22 for 30 and 60 min for qRT-PCR analysis.
(B) and (C) The asr3-1 mutant is more resistant to Psm and Pst infections. Four-week-old wild-type and asr3-1 mutant plants were hand-inoculated with bacterial suspension at a density of 5 × 105 cfu/mL, and bacterial population was quantified at 0, 2, and 4 dpi.
(D) Bacterial growth of avirulent strain Pst avrRpt2.
(E) ASR3 complements asr3-1 mutant for FRK1 gene induction. Ten-day-old seedlings from Col-0 wild type, asr3-1 mutant, and complementation lines C3 and C7 were treated with 100 nM flg22 for 60 min for qRT-PCR analysis.
(F) ASR3 complements asr3-1 mutant in Pst-mediated pathogen infection. Four-week-old plants were spray-inoculated with Pst at 108 cfu/mL, and bacterial counting was performed at 0, 3, and 5 dpi.
(G) flg22-induced ROS burst in the wild type and asr3-1 mutant. Leaf discs from 5-week-old plants were treated with water or 100 nM flg22 over 30 min. The data are shown as means ± se from 24 leaf discs.
(H) flg22-induced MAPK activation in the wild type and asr3-1 mutant. Ten-day-old seedlings were treated with 100 nM flg22 and collected at the indicted time points. MAPK activation was analyzed by immunoblot with α-pERK antibody (top panel), and the protein loading is shown by Ponceau S staining for Rubisco (RBC) (bottom panel).
The data in (A) to (F) are shown as mean ± sd from three independent repeats, and the asterisk indicates a significant difference with a Student’s t test (P < 0.05) when compared with the wild type. The above experiments were repeated three times with similar results.