Despite the paradigm that noncellulosic cell wall polysaccharides are synthesized in the Golgi, compelling evidence suggests that (1,3;1,4)-β-d-glucans are assembled at the plasma membrane.
Abstract
The current dogma for cell wall polysaccharide biosynthesis is that cellulose (and callose) is synthesized at the plasma membrane (PM), whereas matrix phase polysaccharides are assembled in the Golgi apparatus. We provide evidence that (1,3;1,4)-β-d-glucan (mixed-linkage glucan [MLG]) does not conform to this paradigm. We show in various grass (Poaceae) species that MLG-specific antibody labeling is present in the wall but absent over Golgi, suggesting it is assembled at the PM. Antibodies to the MLG synthases, cellulose synthase-like F6 (CSLF6) and CSLH1, located CSLF6 to the endoplasmic reticulum, Golgi, secretory vesicles, and the PM and CSLH1 to the same locations apart from the PM. This pattern was recreated upon expression of VENUS-tagged barley (Hordeum vulgare) CSLF6 and CSLH1 in Nicotiana benthamiana leaves and, consistent with our biochemical analyses of native grass tissues, shown to be catalytically active with CSLF6 and CSLH1 in PM-enriched and PM-depleted membrane fractions, respectively. These data support a PM location for the synthesis of MLG by CSLF6, the predominant enzymatically active isoform. A model is proposed to guide future experimental approaches to dissect the molecular mechanism(s) of MLG assembly.
INTRODUCTION
The primary plant cell wall is a mechanical network of rigid cellulose microfibrils embedded within a reinforced gel-like phase of matrix (noncellulosic and pectic) polysaccharides. It is vital to plant growth and development, as it determines the functional specialization of cells through regulation of their shape, permeability, and mechanical properties. Walls and their constituent polysaccharides, including mixed-linkage glucan (MLG), also have important roles in the agri-food industry and in human health (Collins et al., 2010).
However, in spite of the importance of walls, both in planta and in agro-industrial applications, we know little about the molecular mechanism(s) of the biosynthesis of their major components, the polysaccharides. Polysaccharide biosynthesis is largely attributed to two major classes of enzymes: several large families of polysaccharide synthases (the cellulose synthase [CesA] superfamily, GT2, and the glucan synthase-like family [GSL], GT48), which are integral transmembrane proteins found in both the plasma membrane (PM) and the Golgi apparatus, and multiple families of type II glycosyltransferases (CAZy, www.cazy.org; Lombard et al., 2014) largely found in the Golgi. The CesA superfamily comprises the cellulose synthases (CesAs) and a large family of CELLULOSE SYNTHASE-LIKE (CSL) genes that encode the backbones of a range of matrix phase polysaccharides, including the glucan backbone of xyloglucans (CSLC), mannans (CSLA), and the MLGs (CSLF/H) (Doblin et al., 2010).
Among the CSLF/H gene families, CSLF6 is the dominant gene responsible for the synthesis of the majority of MLG in the walls of vegetative and floral tissues in grasses. It is the most highly expressed CSLF gene in most tissues of barley (Hordeum vulgare), wheat (Triticum aestivum), Brachypodium distachyon, and rice (Oryza sativa), including developing seedling leaf, coleoptiles, and endosperm (Burton et al., 2008; Kimpara et al., 2008; Doblin et al., 2009; Nemeth et al., 2010; Pellny et al., 2012; Vega-Sánchez et al., 2012; Suliman et al., 2013; Trafford et al., 2013; Schreiber et al., 2014). When CSLF6 expression is reduced either by knockdown or knockout via mutation or T-DNA insertion (Tonooka et al., 2009; Nemeth et al., 2010; Taketa et al., 2012; Vega-Sánchez et al., 2012; Hu et al., 2014), a significant reduction in MLG is observed in both vegetative and floral tissues, indicating its gene product is responsible for the synthesis of the majority of MLG in grasses.
The recent crystal structure determination of the bacterial cellulose synthase (BcsA/B) (Morgan et al., 2013, 2014), together with a computational model of the central cytosolic domain of cotton (Gossypium hirsutum) CesA1 (Sethaphong et al., 2013; Slabaugh et al., 2014), has delivered a major breakthrough in our understanding of the molecular mechanism of cellulose biosynthesis and a clear demonstration that there is only a single active site in both bacterial and plant cellulose synthases. The expression of the CSLF/H proteins in a heterologous dicot system (Arabidopsis thaliana) results in the synthesis of MLG, a polysaccharide that contains both β-(1,3)- and β-(1,4)-glucosidic linkages, suggesting that they are both MLG synthases (Burton et al., 2006; Doblin et al., 2009). This then raises many questions about the mechanism(s) of MLG synthesis, including how each of these proteins with a single active site assembles MLG and where in the cell assembly occurs. While cellulose is synthesized at the PM, it is presumed that the matrix phase polysaccharides are assembled in the Golgi. In an earlier study, we reported that an arabinoxylan (AX)-specific antibody labeled both the wall and Golgi cisternae in endosperm and coleoptiles consistent with this generally accepted paradigm. In contrast, the MLG-specific antibody heavily labeled the walls of barley coleoptiles, endosperm, and suspension-cultured cells (SCCs) yet the adjacent Golgi cisternae were unlabeled (Wilson et al., 2006). These observations are consistent with those of Philippe et al. (2006), who found minimal Golgi labeling in wheat endosperm cells, but differ with those reported by Carpita and McCann (2010), who have shown MLG labeling in the PM and Golgi cisternae in developing maize (Zea mays) coleoptiles.
Here, we provide further evidence to show MLG is primarily synthesized at the PM. We generated antibodies to CSLF6 and CSLH1 proteins allowing reexamination of the location of MLG biosynthesis using complementary approaches. Immuno-transmission electron microscopy (TEM) of multiple tissues of various grass species shows that the final destination of the dominant catalytic protein involved in MLG synthesis, CSLF6, is the PM, verifying our observations in Nicotiana benthamiana leaves that a fluorescent CSLF6 fusion protein overlaps with the PM marker At-PIP2A. Interestingly, by both methods, CSLH1 shows a different subcellular location, being predominantly observed in endoplasmic reticulum (ER) and Golgi membranes, but not the PM. These differences in CSLF6 and CSLH1 location were further verified by membrane fractionation experiments. Topology studies indicate that in both proteins, the central region containing the ‘D, D, D, QXXRW’ motif lies in the cytoplasm and thus are oriented similarly to the CesAs. Together with the MLG location studies, we propose that in grasses MLG assembly, unlike other matrix polysaccharides, occurs primarily at the PM.
RESULTS
Immuno-TEM Indicates That MLG Biosynthesis Does Not Conform to the Paradigm of Other Matrix Phase Polysaccharides
Determining the subcellular location of MLG gives valuable insight into the mechanism of its synthesis. The discrepancy in the cellular distribution of MLG in barley and maize outlined above was attributed by Carpita and McCann (2010) to either fixation artifacts or a possible timing issue, where it was proposed that sampled tissues had ceased synthesizing MLG and thus would not contain MLG in the endomembrane system. This prompted us to revisit our original observations that were based on conventional chemical fixation techniques. We therefore subjected numerous tissue types, at different developmental stages, from various grass species, to cryofixation using high-pressure freezing, a fixation method that upholds polysaccharide and protein epitopes while maintaining optimal preservation of cellular structures (Wilson and Bacic, 2012; McDonald, 2014).
Similar to Arabidopsis (Kang, 2010), root tips of barley and wheat consistently delivered the best ultrastructural fixations, far superior in comparison to other grass species and tissues we investigated. The quality of tissue preservation is evidenced by the PM being appressed to the wall, delineation of membranes with a smooth rather than wavy appearance, Golgi stacks with clearly resolved cisternae, and a uniformly electron-dense cytosol rather than condensed and/or electron-lucent regions (Figure 1; Supplemental Figure 1).
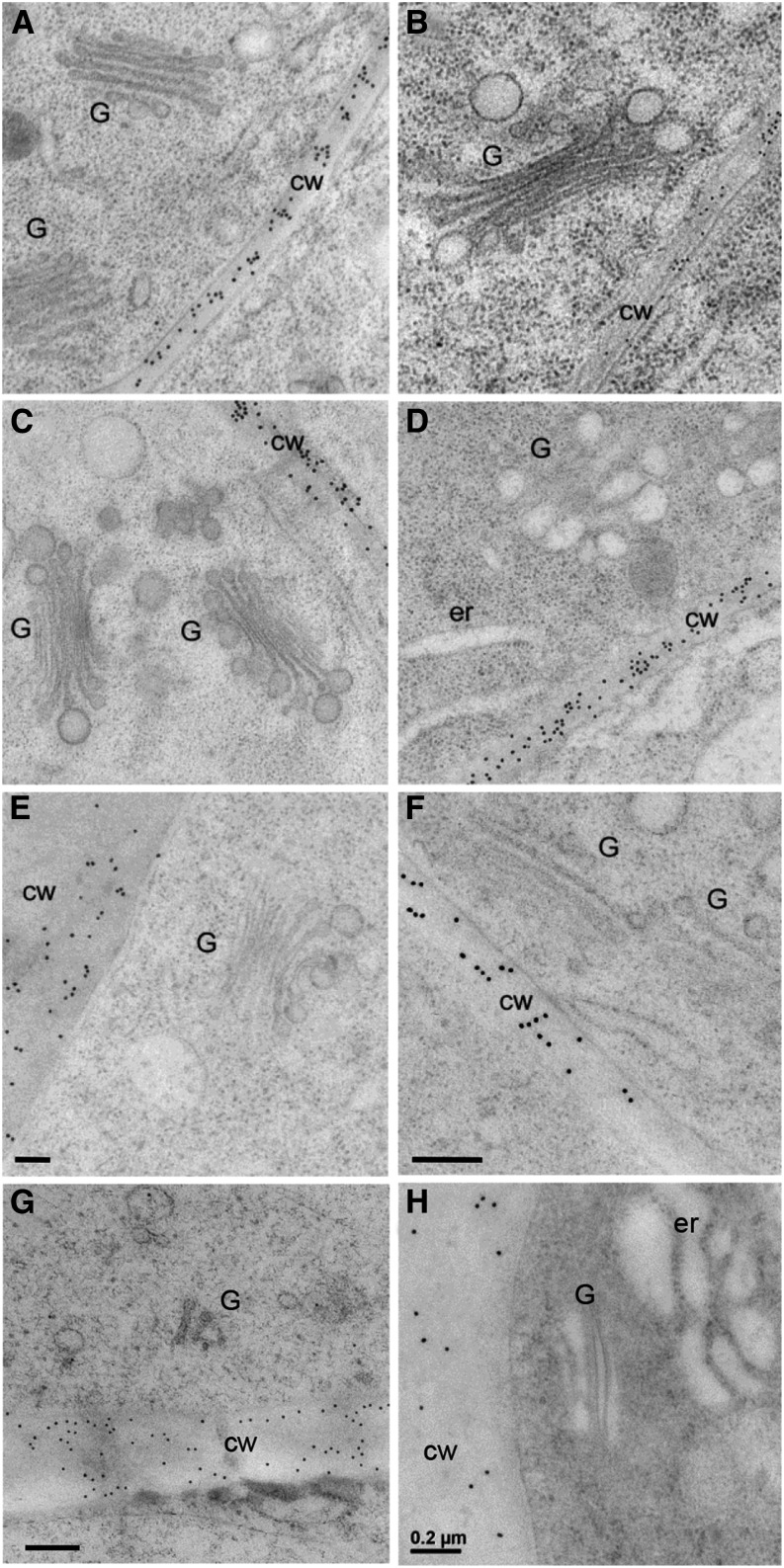
Figure 1.
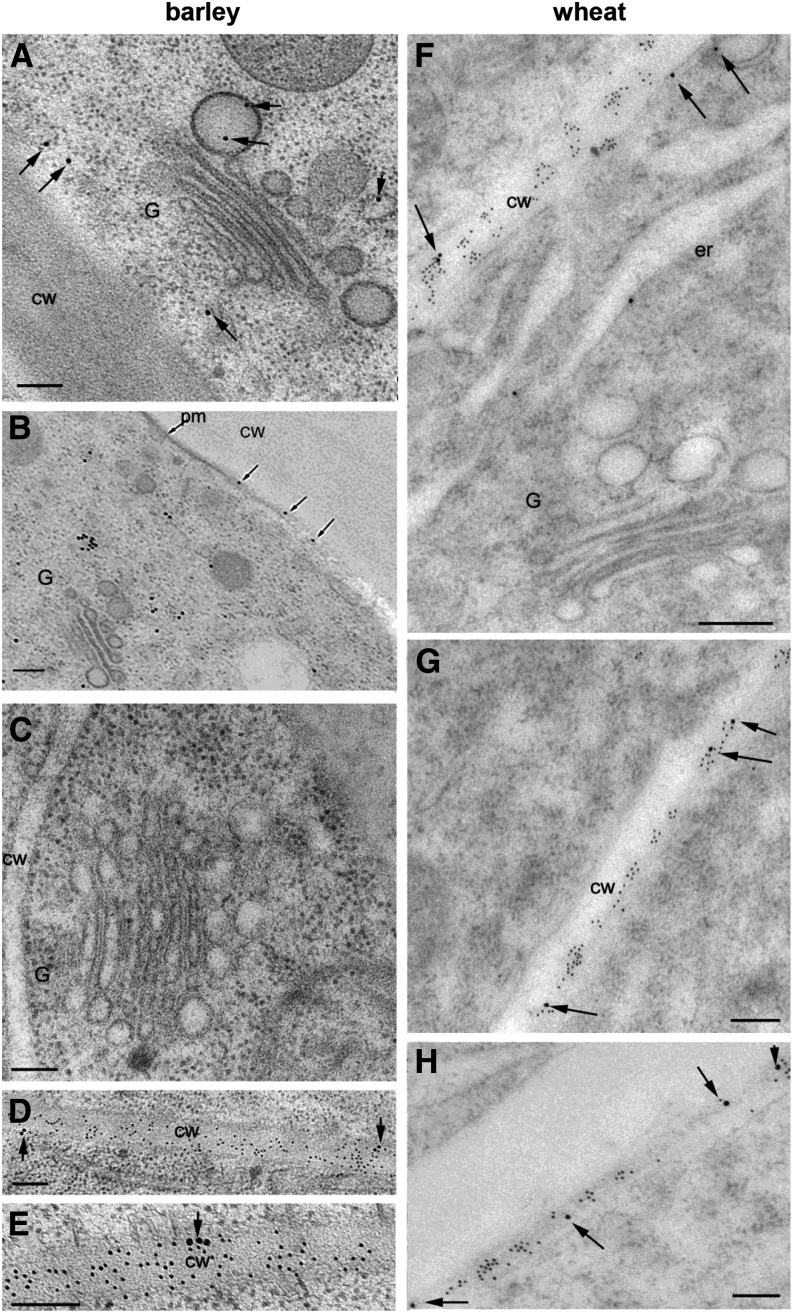
Revealing the Subcellular Location of MLG in Grass Tissues.
MLG is found abundantly along the cell wall in barley (A) and wheat (B) root tip cells but is absent over nearby Golgi. Pretreatment of barley root sections with NaOH (C), proteinase K (D), xylanase (E), or pectinase (F) prior to application of the MLG antibody did not change the labeling pattern. Cell wall labeling is also observed in 7-d-old Lolium SCC (G) and 2-d-old maize coleoptiles but is absent over adjacent Golgi (H). G, Golgi; cw, cell wall; er, endoplasmic reticulum. Bars = 0.2 μm.
As previously reported (Wilson et al., 2006), and observed by others (Trethewey and Harris, 2002), the MLG antibody labels all the walls of cells at the elongation zone in barley and wheat root tips (Figures 1A and 1B, respectively) as well as other vegetative and storage walls of barley, Lolium multiflorum (Italian rye grass), and maize (Figures 1G and 1H; Supplemental Figures 1A to 1C, 1J, and 1K). We also quantified this labeling by counting gold particles in multiple sections in root tissue as well as across a developmental series of barley coleoptiles and in tissues of other grasses where high rates of MLG synthesis occur, such as L. multiflorum suspension-cultured cells (Lolium SCCs) (Figure 2A; Supplemental Table 1). Contrary to the recent report of Carpita and McCann (2010), in none of these cases do we observe any specific labeling above background over the Golgi cisternae with >90% of the labeling present in the cell wall compartment (Figures 1 and 2; Supplemental Figure 1). These observations are consistent across all the Poaceae species we examined and are independent of tissue type. We occasionally observe sporadic labeling of other components of the endomembrane system, such as postsecretory vesicles in transit to the PM and regions of labeling subtending the PM/wall, like those reported by Carpita and McCann (2010), that could be interpreted as reflecting localized regions of MLG biosynthesis prior to deposition into the wall (Figures 1G and 2; Supplemental Figure 1C). However, this pattern for MLG contrasts with the labeling patterns observed for other noncellulosic polysaccharides where labeling in the Golgi is reflective of the intensity in the cell wall (see Supplemental Figures 2A and 2C and description below).
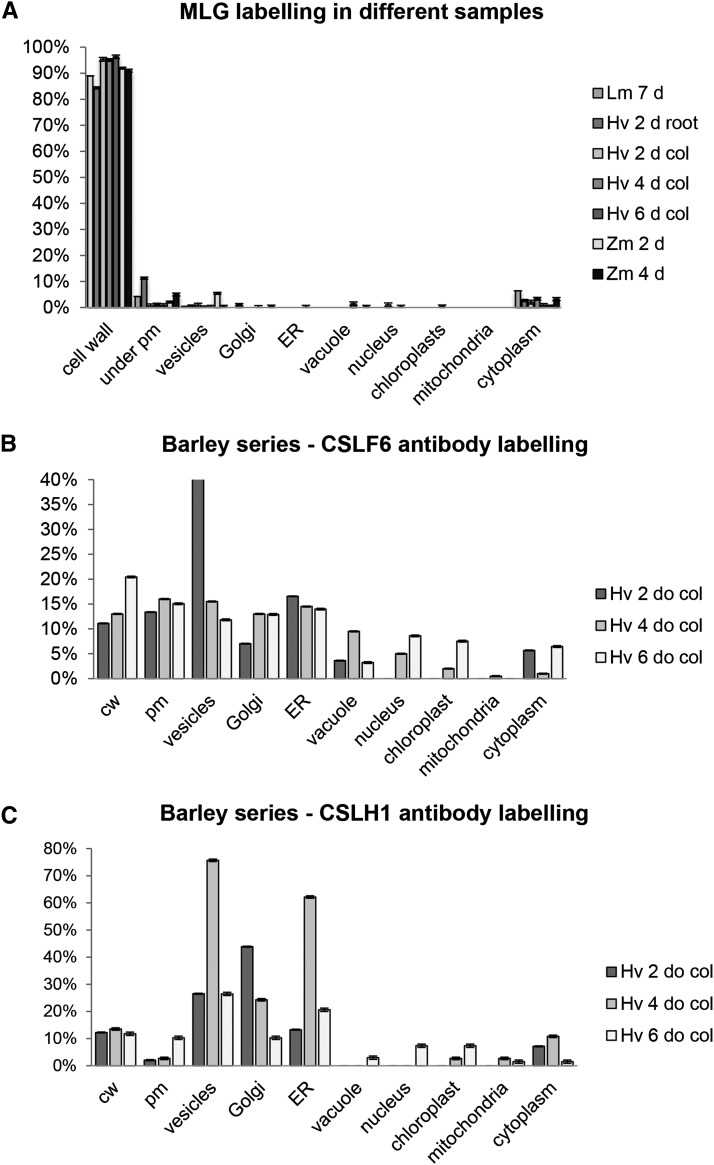
Figure 2.
Percentage of Immunogold Counts in Various Subcellular Locations of MLG, CSLF6, and CSLH.
MLG labeling (A) is shown in a range of tissues and species including Lolium SCC (Lm), barley (Hv) coleoptiles, and root and maize (Zm) coleoptiles. MLG labeling is almost exclusively over the cell wall. Distribution of CSLF6 (B) and CSLH1b (C) antibody labeling in 2-, 4-, and 6-d-old barley coleoptiles. For CSLF6, labeling is found at the PM, whereas there is no binding to PM above background levels for CSLH1.
In Wilson et al. (2006), we proposed that the lack of Golgi labeling by the MLG antibody could possibly be due to the masking of antibody epitopes by nonglycosyl substituents, such as acetyl esters. These are commonly added to matrix phase polysaccharides in the Golgi to increase their solubility and are subsequently removed during deposition of the polysaccharide into the wall (Gille and Pauly, 2012), although there have not been reports of their presence in MLGs (Fincher and Stone, 2004). To test this hypothesis, barley root tip sections were pretreated with alkali (NaOH) to remove ester groups prior to the application of the MLG antibody; no effect on the labeling pattern of MLG (compare Figures 1C [alkali-treated] and 1A [untreated]) was observed. That is, labeling was still observed along the wall but not over the adjacent Golgi. Similarly, pretreatment of sections with proteinase K had no effect on the spatial distribution of MLG labeling (Figure 1D), nor did enzymatic digestion of other polysaccharides by their cognate backbone hydrolases, including AX (Figure 1E) and pectin (Figure 1F). These were tested as AXs are the other major type of matrix polysaccharide in grasses (Doblin et al., 2010), and both xylan (Xue et al., 2013) and pectin (Marcus et al., 2008; Leroux et al., 2011) have been shown to mask the epitopes of other polysaccharides. Together, these observations suggest that MLG epitopes are not present in Golgi (or any other endomembrane compartment) at levels detectable by the MLG-specific antibody, only in (or occasionally subtending) the wall.
In contrast, when AX, xyloglucan, and mannan antibodies were applied to barley root tip sections, labeling was not only seen in walls but also over adjacent Golgi stacks (Supplemental Figures 2A to 2C; Wilson et al., 2006). By comparison, labeling of callose, a noncellulosic polysaccharide known to be synthesized at the PM, was seen associated with plasmodesmata (Supplemental Figure 2D), as expected from previous observations (Brown et al., 1997; Wilson et al., 2006). Thus, our observations with these matrix phase polysaccharide-directed antibodies support the current paradigm of synthesis in Golgi and validate the methodological approaches we employed to investigate MLG biosynthesis. Importantly, our immunocytochemical data lead us to conclude that MLG biosynthesis does not conform to this current paradigm.
CSLF and CSLH Proteins are Membrane-Bound Proteins
A complementary approach to studying the location and/or mechanism of MLG synthesis is to locate their biosynthetic protein catalytic subunits. Since CSLF/H proteins catalyze MLG biosynthesis (see Introduction), we embarked on a program of raising polyclonal antibodies to each of these proteins to use in subcellular location and biochemical experiments (for further details regarding antibody design and characterization, see Supplemental Methods, Supplemental Figures 3 and 4, and Supplemental Data Set 1).
To show experimentally that CSLF6 is a membrane protein, different protein fractions isolated from 4-d-old barley seedlings were probed with anti-CSLF6. A protein band of ∼90 kD was detected almost exclusively in the microsomal membrane (MM) fractions (10,000 to 100,000 g pellet) (Figure 3A). The apparent size of this band is smaller than 105 kD, the theoretical molecular mass of Hv-CSLF6. Anti-CSLF6 appears to bind to the CSLF6 protein in grass species other than barley, as it detects a doublet band of ∼80 to 100 kD in 4- to 7-d-old seedling tissue from wheat, B. distachyon, rice, maize, and L. multiflorum (Figure 3A). A particularly strong signal was also observed in MM isolated from Lolium SCC, a rich source of MLG (Smith and Stone, 1973b) and the model system we are using to purify and characterize the MLG synthase.
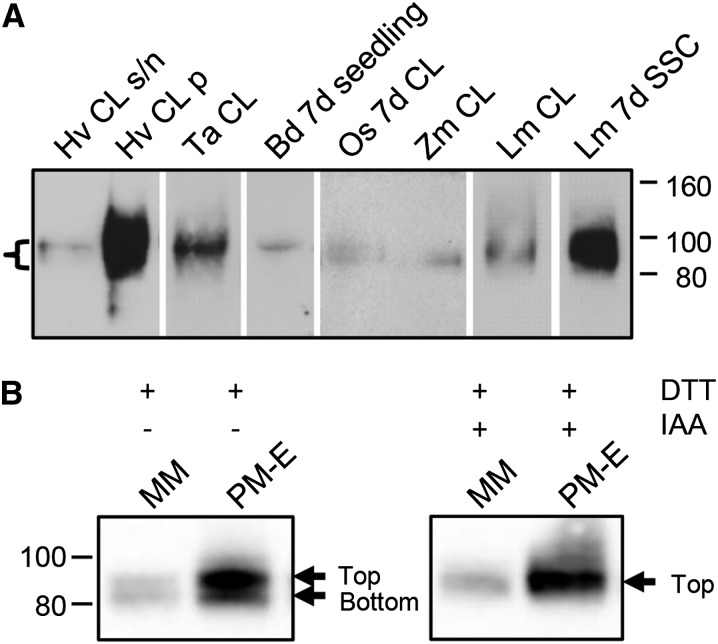
Figure 3.
Immunoblots of Membrane Fractions Prepared from Grass Tissues Using Anti-CSLF6 as a Probe.
(A) MMs prepared from 4-d-old wheat (Ta), B. distachyon (Bd), rice (Os), maize (Zm), and L. multiflorum (Lm) coleoptile and leaf (CL) except for Os sample (7d CL); SSC, 7-d-old Lolium SCC. Supernatant (s/n) and pellet (p, MM) fractions resulting from a 10,000g to 100,000g spin are shown for barley (Hv). Thirty micrograms of MM was loaded per lane except for rice and maize, which contain 40 µg. A band, often observed as a doublet of 80 to 100 kD (bracket), is detected in all grass samples tested. Exposure times and image capture conditions vary between blots. The Benchmark unstained protein ladder was used as the molecular mass marker (in kD).
(B) Immunoblot of Lolium SCC membrane fractions under reducing conditions with and without alkylation prior to SDS-PAGE separation. Upon reduction with DTT, a doublet is normally observed upon immunoblotting using anti-CSLF6 as probe (left panel). The bottom band of the CSLF6 doublet disappears if samples are alkylated with IAA after the DTT reduction step (right panel). PM-enriched fraction (PM-E) generated by PEG-dextran two-phase partitioning.
Detection of a doublet band with anti-CSLF6 raised the possibility that additional CSLF isoforms were also detected by this antibody since multiple CSLF proteins are expressed in seedling leaf, including barley CSLF4, CSLF8, and CSLF10, which have maximal expression in this tissue (Burton et al., 2008). Immunoblotting of MM isolated from N. benthamiana leaves transiently expressing individual barley CSLF isoforms revealed that anti-CSLF6 does not cross-react with other immunodetectable barley isoforms CSLF4 and CSLF10 (Supplemental Methods and Supplemental Figure 5). Thus, we conclude that the doublet is unlikely to be due to isoform cross-reactivity.
To further understand the nature of the CSLF6 doublet, we explored several options, including differences in redox state previously observed for the CesA proteins (Kurek et al., 2002). A portion of the protein samples may either not have been fully reduced upon treatment with DTT or may have been reoxidized during SDS-PAGE analysis. To eliminate the reformation of intra- and/or intermolecular disulphide bonds during electrophoresis, an alkylating agent, iodoacetamide (IAA), was added after the reduction step prior to sample loading. This treatment eliminated the lower band of the doublet detected by anti-CSLF6 both in MM- and PM-enriched fractions (Figure 3B), in keeping with folded proteins having a more compact structure and migrating faster through a gel than unfolded proteins.
Given that the apparent size of the upper CSLF6 band is still 10 to 15 kD smaller than its predicted molecular mass in SDS-PAGE gels, a proteomics approach was used to investigate whether this band represented a full-length version of the CSLF6 protein. When the size of native CSLF6 proteins is compared with heterologously expressed CSLF6, no difference in migration pattern is observed, suggesting that the doublet arises from the full-length protein (Supplemental Figure 6A). Coimmunoprecipitation experiments followed by LC-MSn analyses of tryptic peptides confirmed that in both cases, the doublet is not due to cleavage of a large peptide fragment from either the NH2- or COOH-terminal end of the CSLF6 protein (Supplemental Methods and Supplemental Figures 6B and 6C). Furthermore, we found no evidence in either publically available databases, such as NCBI (http://www.ncbi.nlm.nih.gov/), or the RNA-Seq resources at The James Hutton Institute (http://ics.hutton.ac.uk/morexGenes/) that the molecular mass disparity could result from the alternative CSLF6 transcripts observed in barley. These data reinforce the view that the increased migration of CSLF6 is an inherent feature of this protein, not uncommon to other membrane proteins (Rath et al., 2009), and a phenomenon that has been observed with other CSL proteins (for example, Liepman et al. [2005]).
The pattern and abundance of CSLF6 protein was characterized further in young barley seedlings. Most CSLF6 protein was detected in aerial tissues, particularly in leaf rather than coleoptile, with low levels detected in the root (compare Figures 4A and 4B, left panels). This differential protein abundance is consistent with the relative transcript levels of CSLF6 in these tissues (Burton et al., 2008), providing strong, additional evidence that the protein detected by anti-CSLF6 is indeed CSLF6. In addition, the level of CSLF6 protein in 7-d-old Lolium SCC is equivalent to that observed in barley coleoptile and leaf samples (Figure 4A).
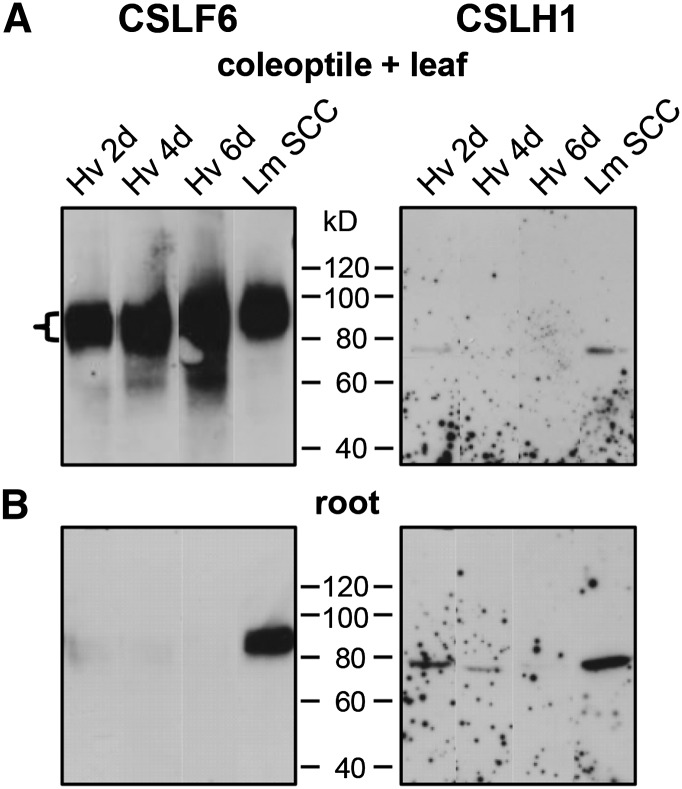
Figure 4.
Immunoblots of MMs Extracted from Barley Seedlings Using Anti-CSLF6 and Anti-CSLHa as Probes.
(A) In MMs (40 µg) extracted from the coleoptile and leaf of 2- to 6-d-old barley seedlings, a strong band of 80 to 100 kD is detected with anti-CSLF6 (left panel). Anti-CSLHa detects a protein of ∼76 kD (right panel). Note the large difference in signal between the CSLF6 and CSLH1 antibodies in these samples. MMs (40 µg) of 7-d-old Lolium SCC were used as a positive control. An overnight exposure is shown for each antibody because this is the length of time required to detect CSLH1 on film.
(B) The bottom panels show matched root samples of the corresponding coleoptile and leaf samples. CSLH1 signal is stronger in root compared with coleoptile and leaf samples.
To show experimentally that CSLH isoforms are also membrane proteins, different protein fractions isolated from barley seedlings were probed with anti-CSLH (for further details of the design and characterization of the CSLH antibodies, see Supplemental Methods, Supplemental Figures 3 and 4, and Supplemental Data Set 1). An ∼76-kD protein was detected by anti-CSLHa in MM of 2- to 6-d-old barley seedling tissues (Figure 4, right panels). Like CSLF6, this protein is smaller in size to the predicted molecular mass of barley CSLH1 at 83 kD. Given steady state transcript levels of CSLH1 are ∼100- to 1000-fold lower than CSLF6 (Burton et al., 2006; Doblin et al., 2009), CSLH1 protein level is, as expected, much lower than CSLF6 (Figure 4, compare left and right panels). Furthermore, the pattern of labeling between the two antibodies in aerial and root tissues is consistent with the pattern expected from the relative transcript abundance of CSLF6 and CSLH1 (Burton et al., 2008; Doblin et al., 2009). Notably, a similar sized band to the barley samples was also detected in Lolium SCC (Figure 4, right panels), indicating anti-CSLHa has cross-reactivity in other grass species and that both CSLF6 and CSLH1 can exist in the same cell type.
In contrast, a protein of ∼65 kD is observed on immunoblots probed with anti-CSLHb (Supplemental Figure 7A, arrows). Like anti-CSLF6 (Figure 3A), anti-CSLHb detects a similar sized protein band in multiple Poaceae species (Supplemental Figure 7B). The apparent size discrepancy of this band with that detected by anti-CSLHa (Figure 4, right panels) was initially thought to be due to the use of different protein molecular mass markers on earlier protein gels (compare Figures 4 and 7); however, this is not the case as a difference in the size of the band detected by the CSLHa and CSLHb antibodies is observed in immunoblots using the same protein molecular mass markers (compare Figures 7 and 9). To confirm that they both detect CSLH1, blots with MMs of N. benthamiana leaves infiltrated with VENUS-Hv-CSLH1 were probed with anti-CSLHa and -CSLHb and shown to detect a band of the same molecular mass that matched the size of the fluorescent VENUS-CSLH1 protein (Supplemental Figures 7C to 7E). While neither antibody has as yet successfully pulled down the native CSLH1 protein in coimmunoprecipitation experiments, we were able to detect the heterologously expressed VENUS-CSLH1 protein using an anti-GFP pull-down approach (Supplemental Figure 7F), verifying that both antibodies bind CSLH1. These data suggest that anti-CSLHa and -CSLHb detect different conformations of the CSLH1 protein. A change in antibody binding pattern after IAA treatment was not observed (Supplemental Figure 7G), suggesting the conformers detected by anti-CSLHa and -CSLHb do not reflect differences in redox state. Further experiments are underway to determine whether CSLH1, like other membrane proteins, runs aberrantly on SDS-PAGE gels due to alterations in detergent binding (Rath et al., 2009).
Figure 7.
Examining the Distribution of CSLF6 and CSLH1 between Intracellular Membranes and PM.
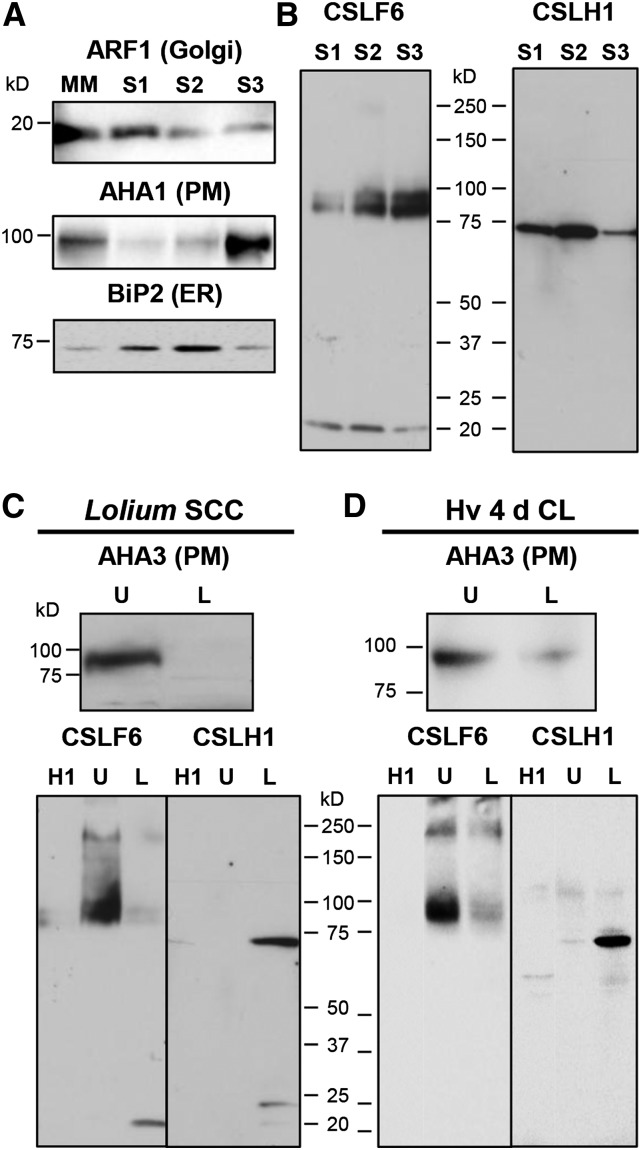
Membranes were isolated using subcellular fractionation with either a step-wise sucrose density gradient (S1, S2, and S3 fractions) ([A] and [B]) or two-phase partitioning (PM-enriched, upper PEG phase [U] and PM-depleted, lower dextran phase [L]) ([C] and [D]). The S1 fraction is enriched in Golgi membranes, whereas the S3 fraction is in PM. ARF1, Golgi marker; H+-ATPase (AHA1 or AHA3), PM marker; BiP2, ER marker. Anti-CSLHa was used as a probe in (B) to (D). The CSLF6 blots shown in (C) and (D) are reprobings after stripping of the same blots probed with anti-CSLHa. Signal was captured electronically in (A) and (D) and on film in (B) and (C).
Figure 9.
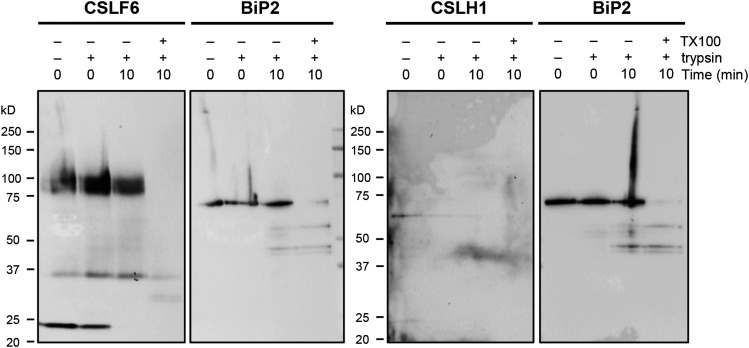
Protease Protection Assay Using MMs Isolated from 7-d-Old Lolium SCC.
MMs prepared from 7-d-old Lolium SCC were digested with trypsin for 10 min before reactions were stopped by addition of trypsin inhibitor. Time zero controls were preincubated with trypsin inhibitor prior to trypsin addition. Detergent-treated samples were preincubated in 0.5% Triton X-100 prior to trypsin digestion. Treated MM aliquots were then run on SDS-PAGE gels, blotted, and probed with either anti-CSLF6 or -CSLHb. Duplicate samples were loaded onto the same SDS-PAGE gel for both CSLF/H and BiP2 blots.
In silico expression analysis of the three rice CSLH genes (www.plexdb.org) (Dash et al., 2012) indicates that these genes are coexpressed in seedlings and, hence, that more than one CSLH isoform is likely to be present in MM extracts of seedling tissues. The theoretical molecular mass range of the rice CSLH isoforms spans ∼5 kD, a large enough difference to be observable on an SDS-PAGE gel (Supplemental Figure 5). As rice CSLH1 transcripts are most abundant of the three genes in seedlings, detection of a single protein suggests that both the CSLH antibodies are CSLH1 specific.
Subcellular Location of CSLF/H Proteins
As a prelude to work in native tissues, we used a fluorescent tag approach in a heterologous system to gain a whole-cell overview of CSLF6 and CSLH1 subcellular location. N. benthamiana leaves were chosen as the heterologous expression system, as this species neither produces MLG nor does its genome contain CSLF and CSLH genes; therefore, the location of the CSLF/H proteins could be studied independently of each other. Expression of VENUS-tagged versions of barley CSLF6 and CSLH1 indicated that these proteins had different subcellular distributions, with the CSLF6 and CSLH1 signals predominantly overlapping the Arabidopsis PM and ER markers PIP2A-CFP and SP-WAK2-CFP-HDEL, respectively (Nelson et al., 2007; Supplemental Figures 8 and 9). However, the highly vacuolated nature of the N. benthamiana leaf epidermal pavement cells and the coarseness of the fluorescent protein signal provided insufficient resolution to confidently assign the location of the CSLF6 and CSLH1 proteins. We therefore performed immuno-TEM and biochemical analyses in native systems to more precisely locate these proteins.
Cryofixed sections of various tissues from a range of Poaceae species, including barley, wheat, Lolium SCC, and maize, were subjected to labeling with anti-CSLF6 and -CSLHa/b. A similar gold particle counting approach was used to quantify the CSLF/H labeling as for the MLG antibody. The pattern of subcellular location with each antibody was found to be consistent across the tissues and species investigated. In all species, anti-CSLF6 labeling was observed in ER, Golgi, post-Golgi secretory vesicles, and the PM (Figures 2B and 5; Supplemental Figures 1D to 1F and Supplemental Table 2). Labeling in these subcellular locations is abolished upon preincubation of anti-CSLF6 with its antigenic peptide (Figure 5C), thereby confirming the binding is to CSLF6. Double labeling shows MLG (10-nm gold) is located in abundance in the wall, while CSLF6 (18-nm gold, arrows) is seen in discrete positions along the PM (Figures 5D to 5H). It should be noted that over time in storage, the CSLF6 antibody displayed an increasing degree of nonspecific binding in immuno-TEM to plastids/nucleus/cell wall that could not be abolished with the antigenic peptide (Figure 2B; Supplemental Table 2).
Figure 5.
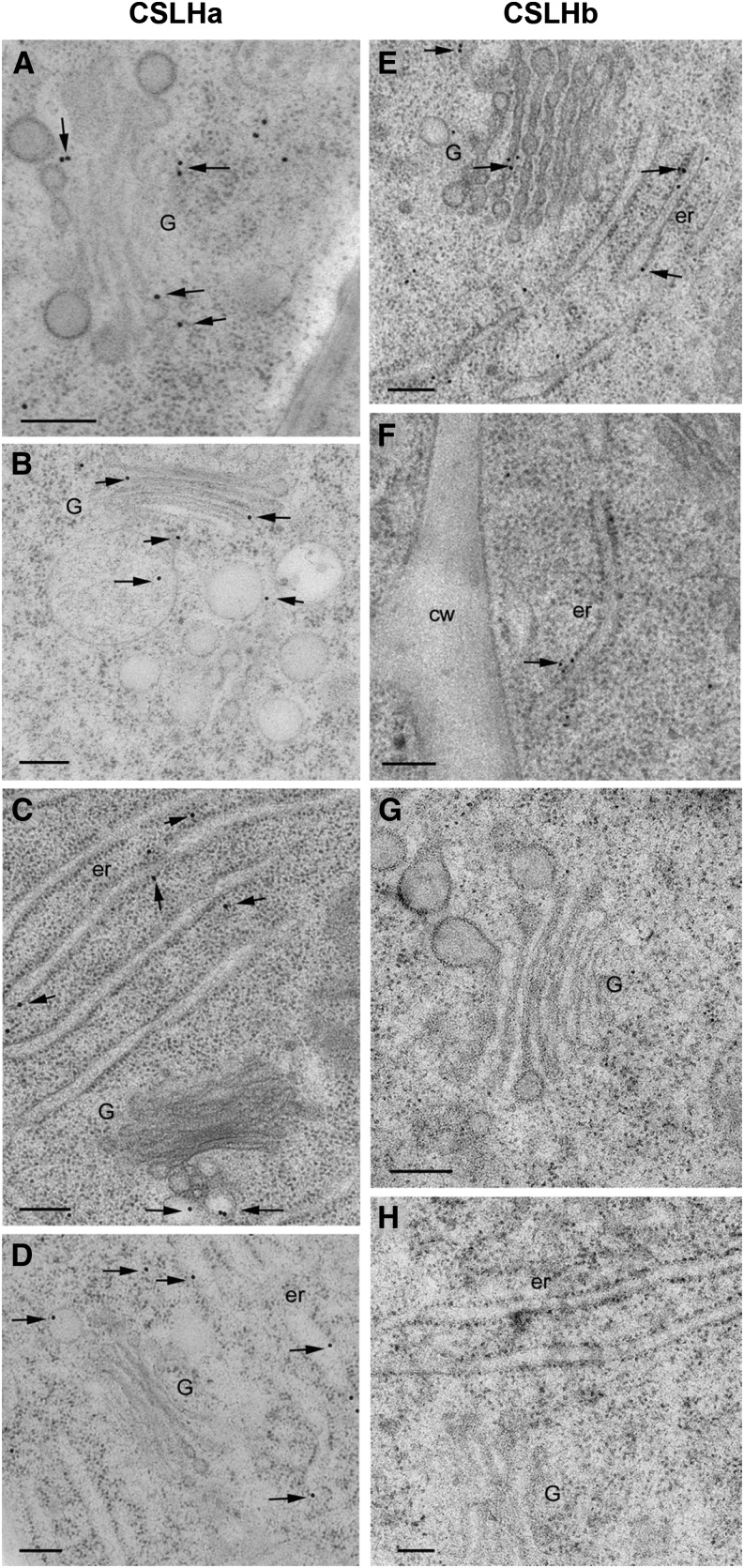
Subcellular Location of CSLF6 in Barley and Wheat Root Tip Cells.
CSLF6 is located in Golgi-derived vesicles and PM in barley ([A], [B], [D], and [E]) and wheat ([F] to [H]). Preincubation of anti-CSLF6 with antigenic peptide 3 (Supplemental Figures 3A and 3B and Supplemental Data Set 1) results in no labeling, as expected (C). Micrographs in (D) to (H) show double labeling with the MLG (10-nm gold) and CSLF6 (18-nm gold, arrows) antibodies. MLG is located abundantly along the wall, while CSLF6 is located in discrete regions along the PM (arrows). G, Golgi; cw, cell wall; pm, plasma membrane. Bars = 0.2 μm.
In contrast, anti-CSLH labeling located CSLH1 to ER, Golgi cisternae, and post-Golgi secretory vesicles in barley (Figures 6A and 6B) and wheat (Figures 6C and 6D) root tip cells using anti-CSLHa. Similar observations were made with anti-CSLHb (Figures 6E and 6F) with labeling being abolished upon preincubation with its antigenic peptide (Figures 6G and 6H). With both CSLH antibodies, we have been unable to detect binding above background levels to the PM in a range of cell types and plant species (Figures 2C and 6F; Supplemental Figures 1G to 1I).
Figure 6.
Comparison of the Subcellular Location of CSLH1 in Barley and Wheat Root Tip Cells Using the Two CSLH Antibodies.
Anti-CSLHa labeling is observed in Golgi, post-Golgi vesicles, and ER in barley ([A], [B], and [E] to [H]) and wheat ([C] and [D]) root tip cells. Application of anti-CSLHb ([E] and [F]) resulted in a similar labeling pattern. Of note, (F) shows ER labeling but no PM labeling. Preincubation of anti-CSLHb with its matching antigenic peptide 6 (Supplemental Figures 3C and 3D and Supplemental Data Set 1) abolishes labeling ([G] and [H]). G, Golgi; cw, cell wall; er, endoplasmic reticulum. Bars = 0.2 μm.
Biochemical Studies Show CSLF6 and CSLH1 Are Also Enriched in Different Subcellular Membrane Fractions
We also adopted a complementary biochemical approach of membrane fractionation to study the location of CSLF6 and CSLH proteins. Initially, we used Lolium SCC as a homogenous source of a single cell type in which both proteins are expressed (Figure 4) and large amounts of material are easy to source. The first method used step-wise sucrose density gradient centrifugation for separation of organelles into three fractions, S1, S2, and S3, respectively. To determine which fraction the organellar membranes partitioned to, antibodies to known marker proteins were used as probes on immunoblots of each membrane fraction including ER (BiP2), Golgi (ADP-ribosylation factor 1 [ARF1]), and PM (anti-AHA1/3, the H+-ATPase). These markers revealed that there is a distribution of membrane types throughout all three fractions but that there is some enrichment of Golgi in S1 (Figure 7A, top panel) and of PM in S3 (Figure 7A, middle panel). When probed with anti-CSLF6, signal intensity was highest in the S3 fraction (Figure 7B, left panel), whereas with anti-CSLHa, the signal was most abundant in the S1 and S2 fractions (Figure 7B, right panel), consistent with our immuno-TEM observations.
As CSLF6 was found in the PM-enriched fraction but contamination by other membranes was significant, we also used a two-phase (polyethylene glycol [PEG]/dextran) partitioning method to obtain highly pure PM from Lolium SCC (Widell et al., 1982; Larsson et al., 1994; Natera et al., 2008). Immunoblot analysis using anti-At-AHA3 (Pardo and Serrano, 1989) showed that the upper PEG phase is enriched in PM proteins and that the lower dextran phase is PM-depleted (Figure 7C). Immunoblotting of the PM-enriched and PM-depleted fractions with anti-CSLF6 and -CSLHa showed that CSLF6 was predominantly located in the PM-enriched fraction (Figure 7C, left panel), whereas the opposite pattern was observed for CSLH1, which was found in the PM-depleted fraction (Figure 7C, right panel). The same observations were made when this PM partitioning method was applied to 4-d-old barley coleoptile and leaf samples (Figure 7D). These data further support the notion that CSLF6 and CSLH1 have different subcellular distributions and final destinations in the cell, with CSLF6 being targeted to the PM, whereas CSLH1 is retained in internal ER/Golgi membranes and is absent from the PM.
The Catalytic Domains of CSLF6 and CSLH1 Are Both Cytoplasmically Oriented
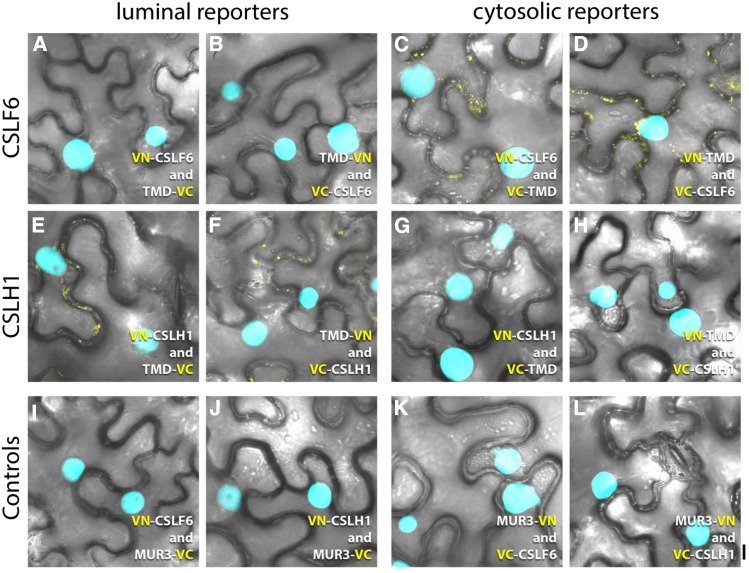
To explore the mechanism of MLG synthesis further, membrane topology of the CSLF6 and CSLH1 proteins was examined using two methods. First, the Golgi Protein Membrane Topology (GO-PROMTO) assay (Søgaard et al., 2012) was used. This system allows the topology of Golgi proteins to be deciphered using a modified bimolecular fluorescence complementation (BiFC) approach and hence is an appropriate choice for CSLF6 and CSLH1 as both proteins locate in part to the Golgi. MUR3 is a type II enzyme with xyloglucan galactosyltransferase activity that is targeted to the Golgi (Madson et al., 2003) and is used as a control in this system. The BiFC test constructs use the first 52 amino acids of the rat sialyltransferase, which contains a transmembrane domain that is sufficient for targeting and retention in the Golgi. The NH2- and COOH-terminal portions of VENUS are fused either before or after the transmembrane domain and hence determine whether they are reporters within the cytosol or Golgi lumen, respectively.
The NH2- and COOH-terminal portions of VENUS (VN and VC, respectively) were fused in frame at the NH2-terminus of CSLF6 and CSLH1, and each protein was coexpressed with the appropriate truncated rat sialyltransferase Golgi luminal (TMD-VN/VC) or cytosolic reporter (VN/VC-TMD) to allow reconstitution of the fluorescent protein. For both genes, each of the four possible pairs of constructs was tested (n ≥ 3). In the case of VN/VC-CSLF6, fluorescence complementation was not observed using the Golgi luminal reporters (Figures 8A and 8B), but signal was consistently detected upon coexpression with the cytosolic reporter (Figures 8C and 8D), indicating its NH2-terminus faces the cytoplasm, as predicted by ARAMEMNON and TOPCONS (Supplemental Figures 3A and 3B). As expected, VN- and VC-CSLF6 combinations with the respective Nicotiana alata MUR3-VC and -VN controls showed no fluorescence complementation (Figures 8I and 8K). In contrast, results for CSLH1 were somewhat equivocal. We observed an inconsistent and very weak fluorescent signal for combinations including VN/VC-CSLH1 with the Golgi luminal reporters (Figures 8E and 8F). An even weaker and more inconsistent signal was seen for combinations including the cytosolic reporter proteins (Figures 8G and 8H), implying the NH2-terminal fusions to CSLH1 had a negative impact on its targeting/folding. No fluorescence was detected with the MUR3 control combinations (Figures 8J and 8L). Thus, it was not possible to unambiguously assign the topology of the NH2-terminus of CSLH1 using the GO-PROMTO assay.
Figure 8.
GO-PROMTO Analysis of Barley CSLF6 and CSLH1 in N. benthamiana Leaves 3 d Postinfiltration.
NH2-terminal VN or VC fusions of CSLF6 ([A] to [D]) and CSLH1 ([E] to [H]) were coexpressed with the Golgi luminal reporters TMD-VC ([A] and [E]) or TMD-VN ([B] and [F]) and the cytosolic reporters VC-TMD ([C] and [G]) or VN-TMD ([D] and [H]), respectively. For CSLF6, the lack of fluorescence complementation with the Golgi luminal reporters ([A] and [B]) but detection of a positive signal with the cytosolic reporters ([C] and [D]) indicates that the NH2-terminus of CSLF6 is located in the cytoplasm. Although weak, a positive signal was detected with the Golgi luminal reporters ([E] and [F]) for CSLH1. However, combinations including CSLH1 and the cytosolic reporters ([G] and [H]) also showed some weak and inconsistent fluorescent signal (arrows). N. alata MUR3-VN and -VC constructs were used as negative controls ([I] to [L]).The nuclear marker CFP-N7 (cyan) was included as a positive transformation control in all experiments. Bar = 10 µm.
Topology prediction tools do not clearly indicate whether the central catalytic domain of each CSL protein lies in the cytosol, particularly for CSLH1 (Supplemental Figure 3). In order to test this hypothesis for CSLF6 directly, additional GO-PROMTO constructs were generated using COOH-terminal truncations of CSLF6. While preliminary experiments located the catalytic domain of CSLF6 and the region between transmembrane helices 6 and 7 (ARAMEMNON prediction; Supplemental Figure 3A) to the cytoplasm, expression was much weaker and variable between transformed cells in comparison to the full-length constructs.
Given these technical hurdles, we then used protease protection assays to confirm the location of the catalytic domain of both CSLF6 and CSLH1. MMs isolated from 7-d-old Lolium SCC were subjected to trypsin treatment either in the presence or absence of the membrane-permeabilizing detergent Triton X-100. The ER luminal marker BiP2 was used as a control. Results from the protease protection assays showed that in the absence of detergent, CSLF6 and CSLH1 were both sensitive to tryptic degradation, as judged by the reduction in signal intensity of the full-length protein band (Figure 9; Supplemental Table 3). Only in the presence of detergent was there substantial degradation of the ER luminal protein, BiP2 (∼78 kD). In the presence of detergent, CSLF6 and CSLH1 degradation was accelerated such that neither protein was detectable after a digestion for 10 min, whereas the full-length BiP protein as well as some degradation products were still evident. The same digestion pattern was observed in the protease protection assays in which either L. multiflorum, barley, or wheat MMs, isolated from 4-d-old coleoptile and leaf samples, were used (Supplemental Figure 10 and Supplemental Table 3). Our interpretation of these data is that while BiP2 is protected from digestion as it lies inside the relatively intact ER membranes, the antigenic regions of CSLF6 and CSLH1, which lie between the conserved D2 and D3 residues of the active site, are not, as their catalytic region lies in the cytoplasm.
Taken together, the GO-PROMTO and protease protection assays indicate that the catalytic domains of CSLF6 and CSLH1 are likely to be localized to the cytoplasm.
Functional Analysis of CSLF6 and CSLH1 Proteins in the N. benthamiana Leaf System
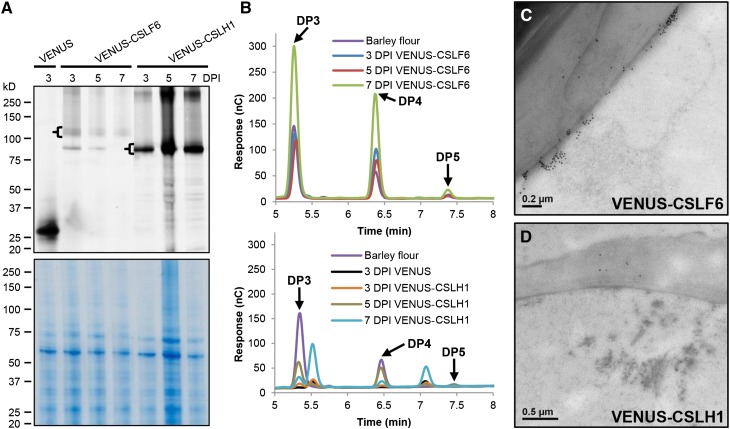
In addition to employing the N. benthamiana leaf system to view the subcellular distribution of CSLF6/H1, we also sought to use it as a functional assay system (Taketa et al., 2012; Vega-Sánchez et al., 2012). Given the abnormal accumulation pattern of VENUS-CSLF6 in large aggregates at the periphery of infiltrated epidermal cells (Supplemental Figure 8), we questioned whether the fluorescently tagged CSLF6 protein was catalytically active. The same leaves imaged by confocal microscopy were harvested and both MMs and walls (alcohol-insoluble residue [AIR]) prepared. Protein gel blot analysis confirmed that the intact ∼125-kD VENUS-CSLF6 fusion protein accumulated to detectable levels in these leaves (Figure 10A). Upon digestion of AIR samples with lichenase, a MLG-specific endohydrolase, the diagnostic oligosaccharides of grass MLGs, the tri- (G4G3GR) and the tetra- (G4G4G3GR) saccharides, were detected (Figure 10B), indicating that the VENUS-tagged CSLF6, like its untagged counterpart (Taketa et al., 2012; Vega-Sánchez et al., 2012), is active in N. benthamiana leaves.
Figure 10.
Functional Testing of VENUS-Tagged CSLF6 and CSLH1 Proteins.
(A) MMs and cell wall samples (AIR) were prepared from N. benthamiana leaves 3, 5, and 7 d postinoculation (DPI) with either VENUS-CSLF6 or VENUS-CSLH1 constructs. Aliquots of MM (10 μg) were run on an SDS-PAGE gel and the fluorescence imaged using a fluorescence imager (upper panel). The tagged versions of CSLF6 and CSLH1 were detected among the population of fluorescent proteins (brackets). The same gel that was imaged for fluorescence was stained with Coomassie blue to show protein loading (lower panel).
(B) AIR samples from (A) were lichenase-digested and analyzed for MLG content using HPAEC. Diagnostic DP3 and DP4 peaks characteristic of MLG were detected in both the VENUS-CSLF6 and VENUS-CSLH1 samples (upper and lower profiles, respectively). The peak adjacent to DP3 is also present, and increases over time, in VENUS samples.
(C) and (D) TEM images of epidermal cells of N. benthamiana leaves (high-pressure frozen at 7 d postinoculation) transformed with either VENUS-CSLF6 (C) or VENUS-CSLH1 (D) and labeled with MLG antibody.
Immuno-TEM was also used to view the location of MLG in the pavement cells of N. benthamiana transformed with the VENUS-tagged CSLF6. Large ectopic deposits of MLG labeling were observed adjacent to the walls (Figure 10C), coincident with the large aggregates of fluorescent labeling seen with VENUS-CSLF6 expression (Supplemental Figures 8A to 8D and 8M to 8P). These observations suggest that MLG is also produced by CSLF6 protein that accumulates in abnormal aggregates at/subtending the PM, resulting in MLG being, in part, aberrantly deposited in N. benthamiana leaves.
Using the same biochemical analyses, the intact VENUS-CSLH1 fusion protein was observed to accumulate postinfiltration (Figure 10A). Interestingly, it accumulated to higher levels and with different kinetics compared with VENUS-CSLF6. However, lichenase treatment of AIR samples generated from VENUS-CSLH1 infiltrated leaves yielded smaller amounts of MLG, as determined by HPAEC-PAD and TEM analysis (Figures 10B and 10D, respectively).
DISCUSSION
On the basis of earlier biochemical investigations associating polysaccharide synthase activities with Golgi-enriched (but not “pure”) membrane fractions, it has been generally accepted that MLG, a prominent matrix phase polysaccharide of the commelinoid monocots, is synthesized in the Golgi as is the case for other noncellulosic polysaccharides such as pectin, xyloglucan, mannan, and AX (Carpita, 1996; Fincher, 2009; Keegstra, 2010). In the case of the latter polysaccharides, an immunocytochemical approach to their detection with polysaccharide-specific antibodies has provided definitive proof of their synthesis in the Golgi (pectin and xyloglucan [Zhang and Staehelin, 1992]; mannan [Pettolino et al., 2001; Wilson et al., 2006]; AX [Philippe et al., 2006; Wilson et al., 2006]). This same approach has also been used to locate the site of MLG biosynthesis but has drawn contrasting conclusions in different Poaceae species.
MLG Assembly Is Distinct from That of Other Matrix-Phase Polysaccharides
Using high-pressure freezing and freeze substitution to optimally preserve plant tissues, we made similar observations in elongating cortical cells of barley root tips to our earlier chemical fixation study (Wilson et al., 2006), with no MLG detectable above background over the Golgi, only in the wall (Figures 1 and 2). Furthermore, we extended these observations to root tip cells of wheat, maize coleoptiles, and Lolium SCC (Figure 1; Supplemental Figures 1J and 1K), suggesting that this is widespread in the grasses. Chemical (alkali) and enzymatic (endohydrolases) treatments of sections to “unmask” cryptic MLG epitopes did not reveal additional epitopes in the endomembrane system (Figures 1B to 1E); hence, these observations must reflect the mechanism of its synthesis. An absence of MLG labeling over Golgi cisternae has been noted by others studying spatio-temporal patterns of wall deposition in wheat endosperm (Philippe et al., 2006), in maternal integumentary cells of rice endosperm (Brown et al., 1997) and in the Charophycean green alga Micrasterias fimbriata in which a MLG, with a similar fine structure to grasses, is deposited in secondary walls (Eder et al., 2008). These data therefore suggest that (1) the mechanism of synthesis and assembly of MLG is common to other Poaceae species and potentially other more distantly related taxa in the plant lineage, and (2) it is distinct from that for other matrix-phase polysaccharides whose assembly is Golgi located.
CSLF6 and CSLH Are Targeted to Different Subcellular Locations
We used multiple experimental approaches to determine the location of the CSLF6 and CSLH1, the catalytic subunits of the MLG synthase (Burton et al., 2006; Doblin et al., 2009; Burton et al., 2011; Taketa et al., 2012; Vega-Sánchez et al., 2012). Intriguingly, the two classes of CSL proteins showed distinct subcellular locations. Fluorescently tagged versions of CSLF6 and CSLH1 (Supplemental Figures 8 and 9), as well as their endogenous forms (Figures 5 and 6; Supplemental Figure 1), were found in different subcellular compartments. CSLH1 appears earlier in the endomembrane pathway, predominantly locating to ER and Golgi but not the PM, while CSLF6 is mostly observed in postsecretory vesicles, at and subtending the PM.
Transient expression of VENUS-tagged fusion proteins in N. benthamiana leaves showed a considerable overlap of fluorescence for CSLF6 and CSLH1 with the PM and ER markers, respectively (Supplemental Figures 8 and 9). Importantly, fluorescence was associated with expression of a full-length fusion protein in each case (Figure 10A). Using an immuno-TEM approach to locate CSLF6 and CSLH1 in native tissues, we were able to confirm that their intracellular location is not significantly altered by translational fusion of VENUS. The labeling observed with anti-CSLF6 and -CSLH1 in barley and wheat root tip sections (Figures 5 and 6, respectively) showed a similar pattern of subcellular location as for their VENUS-tagged fusion protein variants, but notably without the large aberrant aggregates of CSLF6 at the periphery of the cell (Supplemental Figures 8 and 9; see below for discussion). Labeling with anti-CSLH in native tissues was more difficult to detect in comparison to anti-CSLF6, suggestive of a lower overall abundance of this protein in these cell types. This is consistent with the much lower transcript levels of CSLH1 and the minor role it plays in MLG synthesis in grasses.
A biochemical approach using membrane fractionation confirmed the different locations of the CSLF6 and CSLH1. Using both sucrose density gradient fractionation and two-phase partitioning methods, we demonstrated that CSLF6 and CSLH1 largely partition into PM-enriched and PM-depleted fractions, respectively (Figure 7), indicating that these proteins locate differently in the cell. The slightly different fractionation of the Golgi marker ARF1 to CSLH1 in sucrose density gradient fractions is consistent with fluorescence tagging and immuno-TEM analyses showing the majority of CSLH1 protein locates to ER with a minor component observed in Golgi (compare Figures 7A and 7B), as indicated by its identical pattern to the BiP2 ER marker with maximal signal in the S2 fraction. In conclusion, both the cell biological (native and heterologous systems) and biochemical (membrane fractionation) studies confirm that the MLG is only detected in the wall, whereas the catalytic protein subunits, CSLF and CSLH, are located in the PM and Golgi, respectively, as their final locations.
Using N. benthamiana Leaves as a Functional Assay System
Apart from observing the subcellular location of the VENUS-tagged versions of CSLF6 and CSLH1 in the N. benthamiana leaf system, we tested whether they were functionally active. For both CSLF6 and CSLH1, the presence of MLG in walls was confirmed by chemical and immuno-TEM analysis (Figure 10). This is notable as the large, fluorescent CSLF6 aggregates observed in these cells comprise a significant proportion of the total CSLF6 protein (Supplemental Figure 8). These protein aggregates are artifacts of this expression system as they are rarely observed in native tissues, being coincident with the regions of CSLF6 label observed subtending the PM (compare Supplemental Figure 8 with Figure 5 and Supplemental Figures 1D to 1F). We propose that these large PM-associated CSLF6 protein aggregates are a consequence of incomplete fusion of secretory vesicles with the PM, with the host cell potentially not having the capacity to correctly deliver CSLF6 protein to the PM and/or regulate its levels (and/or other regulatory components) at this location in the normal manner. Aberrant phenotypes associated with CSLF6 overexpression have also been identified in barley (Burton et al., 2011) implying this is a common occurrence in CSLF6 overexpression studies. However, while CSLF6 cellular distribution is partially disrupted, it is catalytically active as shown by the large ectopic deposits of MLG observed within these regions (Figure 10C). This suggests that CSLF6 can participate in MLG synthesis prior to reaching the PM upon heterologous expression in N. benthamiana leaves.
Our observations with the VENUS-CSLH1 construct also suggest some expression artifacts that have been noted by others in overexpression systems (Quattrocchio et al., 2013). Despite significantly higher levels of VENUS-CSLH1 expression in N. benthamiana leaf cells compared with VENUS-CSLF6 (Figure 10A), the MLG product does not accumulate to comparable levels (Figure 10B). This finding implies that much of the CSLH1 protein is inactive, likely correlating with the majority of VENUS-CSLH1 being ER associated with only a small portion of active protein located in the Golgi, its final destination. Our BiFC experiments showing the NH2-terminus of CSLH1 residing in both the Golgi lumen and cytoplasm (Figures 8E to 8H) indicates that not all of the CSLH1 protein is folded correctly and hence some of this protein may also be inactive. Our observations of CSLH1 location are similar to our findings in Arabidopsis transgenic lines stably expressing a 3xHA-tagged version of CSLH1 in which the protein was predominantly detected in ER membranes and to a minor extent in Golgi-associated vesicles in cells, with small amounts of MLG (<0.016%) detectable in their walls (Doblin et al., 2009).
Toward a Model of MLG Assembly
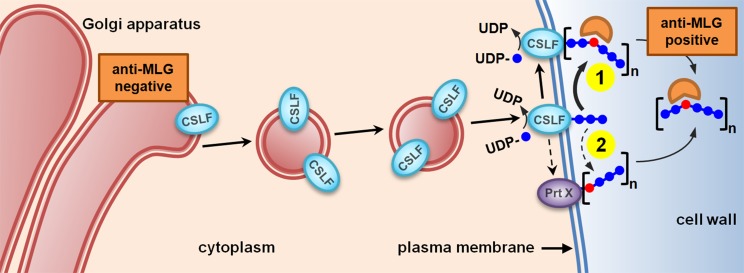
How can the distinctive but overlapping subcellular locations of CSLF6 and CSLH1, along with the absence of MLG in the Golgi be reconciled in a model that explains MLG synthesis? The critical question is whether CSLF and CSLH have the capacity for one or both β-(1,3)- and β-(1,4)-Glc catalytic activities which then dictate the assembly mechanism(s). Figure 11 shows an updated model of MLG synthesis and assembly in grasses first proposed by Doblin et al., (2009) and Burton et al. (2010). The experimental observation that MLG is detected in walls using the MLG-specific antibody is key to formulating a synthesis mechanism. This antibody specifically requires a juxtaposition of both β-(1,3)- and β-(1,4)-glucosidic linkages and a chain length of at least six Glc units for binding; it cannot detect either cello-dextrins [β-(1,4)] or laminari-dextrins [β-(1,3)] (Meikle et al., 1994). Thus, either the MLG is assembled in the Golgi in a form that is inaccessible to the antibody (or below its detection limits) until deposition into the wall or final assembly is at the PM, as occurs for both cellulose and callose. It is important to recall that in vivo CSLF6 is the major MLG synthase of grasses, as indicated by the almost complete lack of MLG in vegetative and reproductive tissues in barley and rice plants lacking a functional CSLF6 (Tonooka et al., 2009; Taketa et al., 2012; Vega-Sánchez et al., 2012).
Figure 11.
Updated Model of the MLG Synthesis Pathway.
CSLF6 is the major MLG synthase and is proposed to act similarly to CesAs, synthesizing MLG chains de novo that are recognized by MLG-specific antibody at the cell surface (scenario 1). A less likely but possible alternative is that CSLF6 produces cello-dextrins (shown here in blue) that are joined together at the plasma membrane by as yet an unidentified protein (Prt X) via a single β-(1,3)-glucosidic linkage (shown in red), creating the MLG chains that are then recognized by the MLG-specific antibody (scenario 2).
“Occam’s razor” principle dictates that the simplest model that fits our data should be chosen, namely, that CSLF6 synthesizes MLG de novo directly at the PM (Figure 11, scenario 1), as occurs with both cellulose and callose synthases. As demonstrated for the bacterial cellulose synthase catalytic subunit BcsA (Morgan et al., 2013; Omadjela et al., 2013) and modeled in the plant CesA equivalent (Sethaphong et al., 2013; Slabaugh et al., 2014), CSLF6 would use UDP-Glc in the cytosol as substrate, synthesize both β-(1,3)- and a β-(1,4)-glucosidic linkages and extrude the nascent β-glucan chain through a channel formed by their own transmembrane helices into the apoplast where it is then detectable by the MLG antibody (Figure 11, scenario 1). This hypothesis is entirely consistent with the activities of GT2 family enzymes (http://www.cazy.org; Lombard et al., 2014) to which CSLF and CSLH proteins belong. This family includes enzymes capable of independently catalyzing the synthesis of either β-(1,3)-glucosidic (e.g., bacterial curdlan synthase, CrdS; Stasinopoulos et al., 1999) or β-(1,4)-glucosidic [e.g., xyloglucan β-(1,4)-glucan synthase, CSLC; Cocuron et al., 2007] linkages. In addition, and of most relevance, is that bifunctional enzymes with one active site capable of synthesizing two types of glycosidic linkages are also included in this family. Examples of these types of GT2 enzymes are the class I streptococcal, vertebrate and viral hyaluronan synthases (Weigel and DeAngelis, 2007), and the galactosyltransferase (GlfT2) that catalyzes the formation of mycobacterial galactan (May et al., 2012). In the former case, two distinct monosaccharides are linked in a disaccharide repeating unit, whereas in the latter, a single monosaccharide is used to generate alternating, regio-isomeric linkages.
In the absence of experimental evidence for this mechanism, we cannot totally exclude other possible mechanisms of MLG assembly no matter how implausible these may be. Thus, if CSLF6 can only synthesize β-(1,4)-glucosidic linkages as has been demonstrated for other CSLs (see above), then cello-oligosaccharides (DP 2 or 3) could be made by CSLF6 either in the Golgi or at the PM and then be joined together via a β-(1,3)-glucosidic linkage at the PM by an as yet unidentified protein (Prt X), creating the MLG glucan chains that are recognized by the MLG-specific antibody (Figure 11, scenario 2). Such an enzyme, presumably an exo- or endotransglycanase that does not conserve the linkage within the acceptor chain, has yet to be identified (Franková and Fry, 2013); hence, this seems an unlikely prospect. However, it is worth noting that transglycosylation of β-glucans within the wall is a common process in yeast/fungi (Latgé, 2010). If such an enzyme did exist, it would need to be present both in grasses and eudicots, given the ability of the Arabidopsis and N. benthamiana systems to synthesize MLG. Recent quantitative trait locus and genome-wide association studies (Islamovic et al., 2013; Shu and Rasmussen, 2014) suggest that other proteins should also be considered to play the role of Prt X in MLG synthesis (and/or regulation) and deserve further investigation.
Although CSLH plays a minor (if any) role in MLG synthesis in the Poaceae (Tonooka et al., 2009; Taketa et al., 2012; Vega-Sánchez et al., 2012), the experimental data indicate a different subcellular mechanism of regulation. CSLH1 is expressed at considerably lower levels than CSLF6 both in vegetative and floral tissues, being transcribed mostly in cells that are undergoing secondary wall thickening, such as interfascicular sclerenchymal fibers and xylem cells (Doblin et al., 2009). Therefore, the simplest explanation of CSLH1 function is that it also catalyzes both β-glucosidic linkages in the MLG chains generated within the Golgi but is regulated differently such that in tissues with high levels of CSLF expression, the CSLH is either inactive or active at levels below the detection limits of the immuno-TEM technique. If this hypothesis is correct, we reasoned that an examination of cell types where CSLH is proposed to have a greater role (see above) including where remnant MLG is detectable in the rice cslf6 mutant (Vega-Sánchez et al., 2012) may reveal internal MLG labeling with the MLG antibody. However, we did not detect a MLG labeling pattern in cells of vascular bundles in the leaf that was distinct from other cell types (Figure 2). A more detailed temporal analysis of these cell types is necessary to be confident MLG is not made intracellularly by CSLH1.
A number of other published observations are consistent with our interpretation of CSLF6 being a bifunctional PM-bound MLG synthase with a different regulatory mechanism from CSLH1. Our interpretation explains why CSLF6 can generate significant amounts of MLG upon heterologous expression in either N. benthamiana (Figure 10B; Taketa et al., 2012; Vega-Sánchez et al., 2012) or Arabidopsis, species that don’t normally produce MLG nor have CSLF genes (Doblin et al., 2009). The proposed role of CSLF6 is also consistent with higher than wild-type MLG levels being present in barley leaves overexpressing CSLF6 to a point where the excess MLG causes “vascular suffocation” (Burton et al., 2011). However, similar plant transformation experiments with other CSLF genes expressed under the same 35S promoter do not result in such high leaf MLG levels, suggesting that there may be inherent differences in the ability of CSLF isoforms to make MLG (Burton et al., 2006, 2011). We are as yet unsure whether these are a consequence of either catalytic ability/function or due to differences related to protein expression (including protein abundance [Supplemental Figure 5], folding, stability, targeting, etc.) and/or interactions with other protein partners that participate in the synthesis/assembly process. While there may also be such variances between CSLH proteins, the observation that the bulk of CSLH protein is retained in the ER both in vivo (Figures 2C and 6; Supplemental Figures 1G to 1I and Supplemental Table 2) and in vitro (Supplemental Figure 9) would suggest that there is an intrinsic difference in their targeting and/or trafficking signal and that this may also have an impact on their ability to participate in MLG synthesis.
The potentially distinct mechanisms of MLG regulation for CSLF and H may be explained by their divergent evolutionary histories. A recent survey of the genomes and transcriptomes of Charophycean green algae and land plants has revealed that CSLH genes have evolved much earlier and independently of CSLF genes, with the former being evident in some ferns and gymnosperms but the latter only appearing later in monocots (Yin et al., 2014).
Despite their different subcellular location, our analysis of CSLF6 and CSLH1 membrane topology suggests that the orientation of the large, central domain containing the catalytic region of these proteins is the same, with the motifs required for substrate binding and catalysis lying in the cytosol (Figure 9). Our data are consistent with the findings of Urbanowicz et al. (2004) showing that in vitro MLG synthesis is significantly reduced in intact vesicles of Golgi-enriched fractions of maize coleoptiles subjected to limited proteolysis without a marked change in activity of IDPase, a Golgi-luminal marker enzyme. Despite the apparent difference in subcellular location of the major MLG synthase in our study, the results of both studies indicate this enzyme is topologically equivalent to cellulose synthase, as is the xyloglucan backbone synthase At-CSLC4 (Davis et al., 2010). Since CSLF and CSLH proteins belong to the same GT2 family, we postulate that they could also form a channel allowing their synthesis products to cross the membrane. However, rigorous empirical testing is required to validate this and other CSL models generated using similar approaches.
Future experimentation will focus on defining the exact molecular steps of MLG synthesis and includes purification of CSLF6 and the identification of putative interacting protein partners that regulate synthesis using coimmunoprecipitation. Furthermore, dynamic molecular modeling of CSLF6 will be used to direct construction of CSLF6 variants that, coupled to in vitro enzyme assays, will provide a powerful tool to study structure-function relationships.
METHODS
Plant Material
Hordeum vulgare cv Flagship and Triticum aestivum cv Chara seed was obtained from Geoff Fincher (ARC Centre of Excellence in Plant Cell Walls, University of Adelaide, Australia). Oryza sativa cv japonica seed was a gracious gift from Alex Johnson (University of Melbourne, Australia). Brachypodium distachyon cv Bd21 seed was obtained from the USDA (Albany, CA; http://brachypodium.pw.usda.gov/), and Lolium multiflorum and maize (Zea mays) seed was sourced from Austra Hort Seed Merchants (http://www.austrahort.com.au/) and Snowy River Seeds, respectively. All grass seeds except rice were imbibed in aerated water for 16 h and sown in containers of water-saturated vermiculite, covered in foil and left for 3 to 7 d at 23°C in a controlled environment growth chamber or glasshouse. Rice grains were imbibed on water-saturated filter paper in Petri dishes and incubated in a growth room under conditions of 12 h day, 28°C/12 h night, 24°C. The Lolium SCC derived from endosperm was maintained in Modified White’s medium as described by Smith and Stone (1973a). Cells in log phase (days 6 to 9) were sampled for various analyses.
TEM and Immunolabeling
Root tips from germinated barley and wheat (∼2 mm) were excised with a razor blade or small clumps of Lolium SCCs were placed in freezer hats containing PBS (137 mM NaCl, 2.7 mM KCl, 10 mM KH2PO4, and 1.8 mM KH2PO4, pH 7.4) and high-pressure frozen (Leica EM PACT2). The frozen samples were then freeze-substituted in a Leica EM AFS unit according to the method of Wilson and Bacic (2012) with 0.1% (w/v) uranyl acetate in acetone as the solvent and Lowicryl HM20 as the embedding resin. The procedures of sectioning, immunolabeling, and staining grids with heavy metals have been described (Wilson and Bacic, 2012). Images were taken using either a Philips BioTwin or FEI Tecnai Spirit transmission electron microscope equipped with a Gatan CCD camera. For the quantification of immunogold labeling, the method from Brownfield et al. (2008) was used.
For epitope masking and CSLF/H binding controls, grids were pretreated in 2 M urea in PBS for 10 min at room temperature before incubation in polysaccharide-specific hydrolases. Endo-1,4-β-xylanase (0.8 units/mL; Megazyme, #E-XYRU6) digestions were conducted in PBS with 0.1% (v/v) Tween 20 for 10 min at room temperature and subsequently washed six times with 2 M urea and 0.1% Tween 20 in PBS. The same procedure was followed for pectinase (0.8 units/mL; Sigma-Aldrich #P-5146) and proteinase K (0.01 mg/mL; Sigma-Aldrich #P-6556) digestions. For alkali treatments, grids containing tissue sections were placed in 1 M NaOH for 30 min and then washed thoroughly with ultra-high-quality (UHQ) water prior to antibody labeling.
Antigenic peptide-antibody binding assays were prepared by incubating CSLF6 and CSLH1 antibodies at concentrations of 1:300 and 1:100, respectively, with their corresponding peptides at 0.5 mg/mL in PBS for 1 h at room temperature on a rotating wheel. Assays were then diluted 1:10 before filtration with an Amicon Ultra 0.5 Centrifugal 3K Filter according to the manufacturer’s instructions. Aliquots of retentate containing peptide-antibody conjugates were applied to urea-treated grids and incubated for 10 min at room temperature followed by six washes as described above. Further immunolabeling steps were as described by Wilson and Bacic (2012).
Antibodies
Monoclonal antibodies specific for arabinoxylan (LM10; McCartney et al., 2005) and xyloglucan (LM15; Marcus et al., 2008) were from PlantProbes. Antibodies to MLG (Meikle et al., 1994), heteromannan (Pettolino et al., 2001), and (1,3)-β-d-glucan (Meikle et al., 1991) were from Biosupplies. A gold-conjugated goat anti-rabbit IgG secondary antibody (10 or 18 nm; Jackson ImmunoResearch) was used at 1:20 for visualization.
Preparation of MMs
Plant tissues were ground using a mortar and pestle and MM prepared according to Doblin et al. (2009) with minor modifications. A low speed spin was conducted at 10,000g prior to the supernatant being layered onto a 1 mL 60% (w/v) sucrose cushion and centrifuged at 100,000g for 1 h at 4°C. MMs were collected at the interface, transferred to a fresh centrifuge tube, and diluted in UHQ water, then repelleted at 100,000g for 1 h at 4°C. The MM pellet was then resuspended in a minimal volume of buffer (50 mM Tris-HCl, pH 7.5, and 1 mM EGTA) prior to use.
Sucrose Step Gradient Membrane Fractionation
Lolium SCCs were homogenized as described above in 40 mL MES buffer (100 mM MES, 2 mM EGTA, and 1 mM EDTA, pH 6.5) containing EDTA-free protease inhibitor cocktail (PIC; one tablet per 50 mL solution; Roche) and 5% (w/v) sucrose. Cell debris was pelleted by centrifugation at 10,000g for 10 min at 4°C. The supernatant was then collected and fractionated using a step-wise sucrose density gradient (20, 35, and 50% [w/v] in MES buffer, 5 mL each gradient) developed by Vincent Bulone and colleagues (KTH, Stockholm, Sweden) by centrifugation at 100,000g for 1 h at 4°C. Proteins/organelles in the S1, S2, and S3 fractions were pelleted at 154,383g for 30 min at 4°C and resuspended in a minimal volume of 100 mM HEPES, pH 7.5, and 1 mM DTT.
PEG/Dextran Two-Phase Partitioning
Enrichment of PM was performed as previously described (Natera et al., 2008) with minor modifications. Lolium SCCs (100 g fresh weight) were homogenized using a mortar and pestle in 200 mL homogenizing buffer (50 mM MOPS, 2 mM EGTA, and 2 mM EDTA, pH7.0) containing PIC. The homogenate was filtered through a double-layer of Miracloth (EMD Millipore) instead of mesh and MMs resuspended in a minimal volume of 5 mM KPO4 buffer, pH 7.5, containing PIC. The resuspended membranes (6 g) were added to an 18-g phase system to produce a 24-g aqueous two-phase system with a final composition of 5.8% (w/w) Dextran T-500 (Pharmacosmos), 5.8% (w/w) PEG-3350 (Sigma-Aldrich), 5 mM KPO4 buffer, pH 7.6, and 5 mM KCl. The phase mixture containing membranes was then processed as described by Larsson et al. (1994). Suspensions of membranes were prepared in a similar manner from the lower phase of the partitioning system.
SDS-PAGE
Protein gel electrophoresis was performed using the NuPAGE Bis-Tris electrophoresis system (Invitrogen) according to the manufacturer’s instructions. Typically, 30 to 40 μg of protein was reduced with 10 mM DTT and 1× NuPAGE sample buffer or a modified version thereof (Doblin et al., 2009). Proteins were separated on Bis-Tris 4 to 12% gradient gels (Novex; Life Technologies) together with a molecular mass marker (Precision Plus Protein Kaleidoscope [Bio-Rad] or Benchmark unstained protein ladder [Life Technologies]). Gels were run with MOPS buffer at 200 V. After electrophoresis, the gel was removed from the cassette and immunoblotted.
Protein Visualization on SDS-PAGE Gels
After electrophoresis, SDS-PAGE gels containing fluorescent protein samples were imaged with a Typhoon 8600 Variable Mode Imager (GE Healthcare) using the following settings: PMT = 600 V, pixel size = 200, scan resolution = 50 dots/cm, and green 532-nm excitation with 526 SP emission filter. After imaging, gels were stained with 0.1% (w/v) Coomassie Brilliant Blue G 250/50% (v/v) methanol/10% (v/v) acetic acid for protein visualization. Gels were destained three times for 30 min with 40% (v/v) ethanol/10% (v/v) acetic acid and then washed in UHQ water before scanning.
Reduction and Alkylation of Protein Samples
Protein samples were reduced with 10 mM DTT and 1× NuPAGE sample buffer at 60°C for 1 h, then immediately alkylated with 50 mM IAA at room temperature for 45 min in the dark. Negative control samples were treated with UHQ water instead of IAA. After incubation, samples were separated by SDS-PAGE as described above.
Immunoblotting
After SDS-PAGE, proteins were blotted onto nitrocellulose membrane (Nitrobind, 0.22 μm; ThermoFisher Scientific) using the XCell II Blot Module (Invitrogen) according to the manufacturer’s protocol. After transfer, blots were processed as described by Doblin et al. (2009). Anti-CSLF6 was used at a 1:100 to 2000 dilution and anti-CSLHa/b at a 1:200 to 750 dilution. For immunoblot detection, either the SuperSignal West Pico or Femto Maximum Sensitivity chemiluminescent substrate (Thermo-Scientific) was used according to the manufacturer’s instructions. Blots were then exposed either to x-ray film (Fuji) or digitally captured using a Chemidoc MP Imaging System (Bio-Rad). X-ray film was processed using an AGFA CP1000 x-ray developer. Protein bands were quantified using Image Lab software v4.1 (Bio-Rad) in autodetection mode. Band detection sensitivity was set to low and band-based background subtraction was enabled (10-mm disc size). The Quantity Tools option was used for relative protein quantitation.
For probing blots containing subcellular membrane fractions, anti-AHA1 (1:2500 dilution of H+-ATPase, AS07 260; Agrisera) and anti-ARF1 (1:1000 dilution, AS08 325; Agrisera) were used to detect the PM and Golgi marker proteins, respectively. The anti-AHA3 (1:8000 dilution of anti-H+-ATPase AHA3 isoform; Pardo and Serrano, 1989) generously provided by Ramon Serrano (Universidad Politecnica de Valencia-CSIC, Valencia, Spain) was used as an alternative. For probing protease protection assay blots, anti-BiP2 (Agrisera AS09 481) was used at a 1:2000 dilution.
To allow reprobing with a different antibody, blots were incubated in stripping solution (50 mM Tris-HCl, 2% [w/v] SDS, and 50 mM DTT, pH 7) at 37°C for 15 min, followed by three washes for 30 min each in TBST (0.05% [v/v] Tween 20 in TBS). The blot was then incubated in blocking solution overnight before being reprobed.
Generation of Subcellular Localization Constructs
The 35S:VENUS-CSLF6 and 35S:VENUS-CSLH1 constructs were generated using the GeneArt Seamless Cloning Kit (Life Technologies; #A13288). Briefly, the yellow fluorescent protein variant VENUS (Nagai et al., 2002) was translationally fused upstream of an eight-amino acid linker sequence and the coding sequence of barley CSLF6 or CSLH1 and cloned into a linearized (KpnI and BamHI) pART7 backbone (Gleave, 1992) containing the 35S promoter and the 3′ OCS terminator sequence (see Supplemental Table 4 for primer sequences). The expression cassette was then removed by NotI digestion and inserted into the binary vector pMLBART (Gleave, 1992). Sequence-verified plasmids were subsequently transformed into Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991) by electroporation and colonies PCR verified as previously described (Lampugnani et al., 2012).
Transient Expression in Nicotiana benthamiana Leaves
Constructs were either singly or cotransformed with the ER, Golgi, and PM organelle marker constructs described by Nelson et al. (2007) into N. benthamiana leaves according to the method outlined by Kaplan-Levy et al. (2014) and leaf tissue collected 3 d postinfiltration for analysis. The organelle markers are all COOH-terminal translational fusions between the cyan fluorescent protein variant mCERULEAN (Rizzo et al., 2004) and either At-PIP2A (PM), the first 49 amino acids of Gm-Man1, soybean (Glycine max) α-(1,2)-mannosidase I (Golgi), or the combination of the At-WAK2 signal peptide and the HDEL ER retention signal at the NH2- and COOH-terminus of CFP, respectively (Nelson et al., 2007). All markers were sourced from the ABRC (PM-CK, CD3-1001; G-CK, CD3-961; ER-CK, CD3-953).
Fluorescence Microscopy
Transiently transformed N. benthamiana leaf samples were excised and initially screened for intensity on a Leica MZFLIII dissecting fluorescence microscope, and 0.5-cm2 sections were mounted on a glass slide in UHQ water and then viewed on an inverted Leica SP2 confocal microscope using a 63× PL Apo BL oil objective (numerical aperture of 1.4). A 405-nm laser line, attenuated to 30%, was used to excite mCERULEAN, while a 514-nm laser line, attenuated to 20%, was sequentially used to excite VENUS. Emissions were detected between 415 and 480 nm and 498 and 530 nm, and photodetectors were set at 700, offset by −5. All settings were held constant. Images were collected using the average of eight optical slices.
GO-PROMTO Analysis
The GO-PROMTO assay (Søgaard et al., 2012) was used to test membrane protein topology with some modifications. To improve the signal-to-noise ratio and lower the likelihood of self-assembly, the VENUS protein was split at amino acid 155 and amino acid 152 changed from Ile to Leu (Kodama and Hu, 2010). Genes of interest were cloned in frame with VN and VC in a modified pGREEN II backbone (Hellens et al., 2000) using the GeneArt Seamless Cloning technique (Life Technologies) and transformed into Agrobacterium strain AGL1 for transient expression as described above. The nuclear marker CFP-N7 (cyan) (Kaplan-Levy et al., 2014) was a generous gift from Tezz Quon (Monash University, Melbourne, Australia) and was infiltrated with all combinations as a positive transformation control.
Protease Protection Assay
MMs were prepared from fresh 7-d-old Lolium SCCs as described above with minor modifications. Insoluble polyvinylpolypyrrolidone (0.05% [w/v]) was added to the homogenization buffer prior to grinding in a mortar and pestle. Membranes were pelleted directly rather than being collected on a sucrose cushion and resuspended in a minimal volume of 50 mM Tris-HCl, pH 9.0, and 0.5 M sucrose.
A total of 50 µg of MM protein for CSLF6 and CSLH1 was assayed at a final concentration of 0.6 µg/μL in 50 mM Tris-HCl, pH 9, 20 mM MgCl2, 8 mM CaCl2, and 0.2 M sucrose. Assays were digested with a trypsin (TPCK treated; Sigma-Aldrich #T-8642) to protein ratio of 1:50 (CSLF6) or 1:200 (CSLH1) for up to 10 min in a thermomixer at 23°C, 300 rpm. Digests were stopped with 2 µg/μL Pefabloc SC PLUS, a serine protease inhibitor, including PSC-Protector solution (Roche Applied Science) and incubated for a further 20 min at 23°C. For time 0 digest controls, the protease was preincubated with 2 µg/μL Pefabloc SC in assay buffer for 20 min prior to MM addition and incubated for a further 20 min. Detergent-treated samples were preincubated with 0.5% (v/v) Triton X-100 (Sigma-Aldrich T-8787) for 30 min on ice prior to the addition of protease. Identical aliquots of each assay were loaded onto duplicate SDS-PAGE gels and immunoblotted as described above.
Cell Wall Preparation and MLG Analyses
Preparation of wall material (as an AIR) from N. benthamiana leaves was as described by Doblin et al. (2009). AIR (10 to 15 mg per sample) was analyzed for MLG content by (1,3;1,4)-β-d-glucan endohydrolase (lichenase) digestion and subsequent detection of digestion products by HPAEC as described by Doblin et al. (2009).
Accession Numbers
Sequence data used in this article can be found in the GenBank/EMBL database under the following accession numbers: Hv-CSLF6 (EU267181), Hv-CSLH1 (FJ459581), Os-CSLF6 (LOC_Os08g06380.1), and Os-CSLH1 (LOC_Os10g20090.1).
Supplemental Data
Supplemental Figure 1. Immunogold Labeling of a Barley and Maize Coleoptile Developmental Series with MLG, CSLF6, and CSLHb Antibodies.
Supplemental Figure 2. Revealing the Subcellular Location of Noncellulosic Polysaccharides in 3-d-Old Barley Root Tip Cells by Applying Antibodies Specific to Cell Wall Polysaccharides.
Supplemental Figure 3. Predicted Topology of the CSLF6 and CSLH1 Proteins.
Supplemental Figure 4. CSLF6 and CSLH Antigenic Peptide Dot Blots.
Supplemental Figure 5. Testing the Cross-Reactivity of Anti-CSLF6 to Other Barley CSLF Isoforms.
Supplemental Figure 6. The Smaller Than Predicted Size of CSLF6 on SDS-PAGE Gels Is Unlikely to Be Due to Protein Cleavage.
Supplemental Figure 7. Characterization of the CSLHb Polyclonal Antibody.
Supplemental Figure 8. Transient Heterologous Expression of 35S:VENUS-CSLF6 in N. benthamiana Leaves.
Supplemental Figure 9. Transient Heterologous Expression of 35S:VENUS-CSLH1 in N. benthamiana Leaves.
Supplemental Figure 10. Protease Protection Assays Using MM Isolated from the Aerial Parts of Lolium, Barley, and Wheat Seedlings.
Supplemental Table 1. Percent Immunogold Counts in Cells of Various Grass Species and Tissues Using the MLG Antibody.
Supplemental Table 2. Percent Immunogold Counts in 2-, 4-, and 6-d-Old Barley Coleoptiles Using the CSLF6 and CSLH Antibodies.
Supplemental Table 3. Quantification of Full-Length CSLF6, CSLH1, and BiP2 Bands Detected in Protease Protection Assays.
Supplemental Table 4. Primers Used in Cloning.
Supplemental Data Set 1. CSLF6 and CSLH1 Antigenic Peptides.
Supplementary Material
Acknowledgments
We thank Cherie Beahan and Marija Todorovic-Glavanic for their technical assistance and Rachel Burton for the T7-tagged CSLF/H constructs. We acknowledge the support of the Australian Research Council for funding to the ARC Centre of Excellence in Plant Cell Walls, a Discovery Project grant (DP110100410), and the Commonwealth Scientific and Research Organization Flagship Collaborative Research Program, provided to the High Fibre Grains Cluster via the Food Futures Flagship. Y.Y.H. acknowledges the support of a Melbourne Research Scholarship.
AUTHOR CONTRIBUTIONS
S.M.W., Y.Y.H., E.R.L., A.M.L.V.d.M., M.B., and M.S.D. conducted the experiments and analyzed the data. M.S.D. and A.B. supervised the work. A.B. and M.S.D. conceived the study. S.M.W., M.S.D., and A.B. wrote the article.
Glossary
- MLG
mixed-linkage glucan
- PM
plasma membrane
- AX
arabinoxylan
- SCC
suspension-cultured cell
- TEM
transmission electron microscopy
- ER
endoplasmic reticulum
- IAA
iodoacetamide
- MM
microsomal membrane
- PEG
polyethylene glycol
- GO-PROMTO
Golgi Protein Membrane Topology
- BiFC
bimolecular fluorescence complementation
- AIR
alcohol-insoluble residue
- UHQ
ultra-high-quality
- PIC
protease inhibitor cocktail
Footnotes
Articles can be viewed online without a subscription.
References
- Brown R.C., Lemmon B.E., Stone B.A., Olsen O.A. (1997). Cell wall (1→3)- and (1→3, 1→4)-beta-glucans during early grain development in rice (Oryza sativa L.). Planta 202: 414–426. [DOI] [PubMed] [Google Scholar]
- Brownfield L., Wilson S., Newbigin E., Bacic A., Read S. (2008). Molecular control of the glucan synthase-like protein NaGSL1 and callose synthesis during growth of Nicotiana alata pollen tubes. Biochem. J. 414: 43–52. [DOI] [PubMed] [Google Scholar]
- Burton R.A., Gidley M.J., Fincher G.B. (2010). Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 6: 724–732. [DOI] [PubMed] [Google Scholar]
- Burton R.A., Jobling S.A., Harvey A.J., Shirley N.J., Mather D.E., Bacic A., Fincher G.B. (2008). The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 146: 1821–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R.A., Wilson S.M., Hrmova M., Harvey A.J., Shirley N.J., Medhurst A., Stone B.A., Newbigin E.J., Bacic A., Fincher G.B. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311: 1940–1942. [DOI] [PubMed] [Google Scholar]
- Burton R.A., et al. (2011). Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnol. J. 9: 117–135. [DOI] [PubMed] [Google Scholar]
- Carpita N.C. (1996). Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 445–476. [DOI] [PubMed] [Google Scholar]
- Carpita N.C., McCann M.C. (2010). The maize mixed-linkage (1->3),(1->4)-β-D-glucan polysaccharide is synthesized at the Golgi membrane. Plant Physiol. 153: 1362–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J.-C., Lerouxel O., Drakakaki G., Alonso A.P., Liepman A.H., Keegstra K., Raikhel N., Wilkerson C.G. (2007). A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA 104: 8550–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins H.M., Burton R.A., Topping D.L., Liao M.-L., Bacic A., Fincher G.B. (2010). Variability in fine structures of noncellulosic cell wall polysaccharides from cereal grains: Potential importance in human health and nutrition. Cereal Chem. 87: 272–282. [Google Scholar]
- Dash S., Van Hemert J., Hong L., Wise R.P., Dickerson J.A. (2012). PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 40: D1194–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J., Brandizzi F., Liepman A.H., Keegstra K. (2010). Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 64: 1028–1037. [DOI] [PubMed] [Google Scholar]
- Doblin M., Pettolino F., Bacic A. (2010). Plant cell walls: the skeleton of the plant world. Funct. Plant Biol. 37: 357–381. [Google Scholar]
- Doblin M.S., Pettolino F.A., Wilson S.M., Campbell R., Burton R.A., Fincher G.B., Newbigin E., Bacic A. (2009). A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5996–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder M., Tenhaken R., Driouich A., Lütz-Meindl U. (2008). Occurrence and characterization of arabinogalactan-like proteins and hemicelluloses in Micrasterias (Streptophyta). J. Phycol. 44: 1221–1234. [DOI] [PubMed] [Google Scholar]
- Fincher G.B. (2009). Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 149: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher G.B., Stone B.A. (2004). Chemistry of nonstarch polysaccharides. In Encyclopedia of Grain Science, Wrigley C., Corke H., Walker C.E., eds (Oxford, UK: Elsevier; ), pp. 206–223. [Google Scholar]
- Franková L., Fry S.C. (2013). Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. J. Exp. Bot. 64: 3519–3550. [DOI] [PubMed] [Google Scholar]
- Gille S., Pauly M. (2012). O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hu G., Burton C., Hong Z., Jackson E. (2014). A mutation of the cellulose-synthase-like (CslF6) gene in barley (Hordeum vulgare L.) partially affects the β-glucan content in grains. J. Cereal Sci. 59: 189–195. [Google Scholar]
- Islamovic E., Obert D.E., Oliver R.E., Harrison S.A., Ibrahim A., Marshall J.M., Miclaus K.J., Hu G., Jackson E.W. (2013). Genetic dissection of grain beta-glucan and amylose content in barley (Hordeum vulgare L.). Mol. Breeding 31: 15–25. [Google Scholar]
- Kang B.-H. (2010). Electron microscopy and high-pressure freezing of Arabidopsis. In Methods in Cell Biology, Thomas M.-R., ed (Burlington, MA: Academic Press; ), pp. 259–283. [DOI] [PubMed] [Google Scholar]
- Kaplan-Levy R.N., Quon T., O’Brien M., Sappl P.G., Smyth D.R. (2014). Functional domains of the PETAL LOSS protein, a trihelix transcription factor that represses regional growth in Arabidopsis thaliana. Plant J. 79: 477–491. [DOI] [PubMed] [Google Scholar]
- Keegstra K. (2010). Plant cell walls. Plant Physiol. 154: 483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpara T., Aohara T., Soga K., Wakabayashi K., Hoson T., Tsumuraya Y., Kotake T. (2008). β-1,3:1,4-Glucan synthase activity in rice seedlings under water. Ann. Bot. (Lond.) 102: 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Hu C.-D. (2010). An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques 49: 793–805. [DOI] [PubMed] [Google Scholar]
- Kurek I., Kawagoe Y., Jacob-Wilk D., Doblin M., Delmer D. (2002). Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc. Natl. Acad. Sci. USA 99: 11109–11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani E.R., Kilinc A., Smyth D.R. (2012). PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J. 71: 724–735. [DOI] [PubMed] [Google Scholar]
- Larsson C., Sommarin M., Widell S., Harry W., Gote J. (1994). Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. In Methods in Enzymology, H. Walter and G. Johansson, eds (San Diego, CA: Academic Press; ), pp. 451–469. [Google Scholar]
- Latgé J.-P. (2010). Tasting the fungal cell wall. Cell. Microbiol. 12: 863–872. [DOI] [PubMed] [Google Scholar]
- Lazo G.R., Stein P.A., Ludwig R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (NY) 9: 963–967. [DOI] [PubMed] [Google Scholar]
- Leroux O., Knox J.P., Masschaele B., Bagniewska-Zadworna A., Marcus S.E., Claeys M., van Hoorebeke L., Viane R.L.L. (2011). An extensin-rich matrix lines the carinal canals in Equisetum ramosissimum, which may function as water-conducting channels. Ann. Bot. (Lond.) 108: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A.H., Wilkerson C.G., Keegstra K. (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA 102: 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42: D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M., Dunand C., Li X., Verma R., Vanzin G.F., Caplan J., Shoue D.A., Carpita N.C., Reiter W.-D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J.J., Farkas V., Pedersen H.L., Willats W.G.T., Knox J.P. (2008). Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May J.F., Levengood M.R., Splain R.A., Brown C.D., Kiessling L.L. (2012). A processive carbohydrate polymerase that mediates bifunctional catalysis using a single active site. Biochemistry 51: 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L., Marcus S.E., Knox J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53: 543–546. [DOI] [PubMed] [Google Scholar]
- McDonald K.L. (2014). Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma 251: 429–448. [DOI] [PubMed] [Google Scholar]
- Meikle P.J., Bonig I., Hoogenraad N.J., Clarke A.E., Stone B.A. (1991). The location of (1→3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-β-glucan-specific monoclonal antibody. Planta 185: 1–8. [DOI] [PubMed] [Google Scholar]
- Meikle P.J., Hoogenraad N.J., Bonig I., Clarke A.E., Stone B.A. (1994). A (1→3,1→4)-β-glucan-specific monoclonal antibody and its use in the quantitation and immunocytochemical location of (1→3,1→4)-β-glucans. Plant J. 5: 1–9. [DOI] [PubMed] [Google Scholar]
- Morgan J.L.W., Strumillo J., Zimmer J. (2013). Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J.L.W., McNamara J.T., Zimmer J. (2014). Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20: 87–90. [DOI] [PubMed] [Google Scholar]
- Natera S.H.A., Ford K.L., Cassin A.M., Patterson J.H., Newbigin E.J., Bacic A. (2008). Analysis of the Oryza sativa plasma membrane proteome using combined protein and peptide fractionation approaches in conjunction with mass spectrometry. J. Proteome Res. 7: 1159–1187. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nemeth C., et al. (2010). Down-regulation of the CSLF6 gene results in decreased (1,3;1,4)-β-D-glucan in endosperm of wheat. Plant Physiol. 152: 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omadjela O., Narahari A., Strumillo J., Mélida H., Mazur O., Bulone V., Zimmer J. (2013). BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc. Natl. Acad. Sci. USA 110: 17856–17861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J.M., Serrano R. (1989). Structure of a plasma membrane H+-ATPase gene from the plant Arabidopsis thaliana. J. Biol. Chem. 264: 8557–8562. [PubMed] [Google Scholar]
- Pellny T.K., Lovegrove A., Freeman J., Tosi P., Love C.G., Knox J.P., Shewry P.R., Mitchell R.A.C. (2012). Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-Seq transcriptome. Plant Physiol. 158: 612–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino F.A., Hoogenraad N.J., Ferguson C., Bacic A., Johnson E., Stone B.A. (2001). A (1—>4)-beta-mannan-specific monoclonal antibody and its use in the immunocytochemical location of galactomannans. Planta 214: 235–242. [DOI] [PubMed] [Google Scholar]
- Philippe S., Saulnier L., Guillon F. (2006). Arabinoxylan and (1→3),(1→4)-β-glucan deposition in cell walls during wheat endosperm development. Planta 224: 449–461. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F.M., Spelt C., Koes R. (2013). Transgenes and protein localization: myths and legends. Trends Plant Sci. 18: 473–476. [DOI] [PubMed] [Google Scholar]
- Rath A., Glibowicka M., Nadeau V.G., Chen G., Deber C.M. (2009). Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 106: 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo M.A., Springer G.H., Granada B., Piston D.W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22: 445–449. [DOI] [PubMed] [Google Scholar]
- Schreiber M., Wright F., MacKenzie K., Hedley P.E., Schwerdt J.G., Little A., Burton R.A., Fincher G.B., Marshall D., Waugh R., Halpin C. (2014). The barley genome sequence assembly reveals three additional members of the CslF (1,3;1,4)-β-glucan synthase gene family. PLoS ONE 9: e90888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethaphong L., Haigler C.H., Kubicki J.D., Zimmer J., Bonetta D., DeBolt S., Yingling Y.G. (2013). Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. USA 110: 7512–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X., Rasmussen S.K. (2014). Quantification of amylose, amylopectin, and β-glucan in search for genes controlling the three major quality traits in barley by genome-wide association studies. Front. Plant Sci 5: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh E., Davis J.K., Haigler C.H., Yingling Y.G., Zimmer J. (2014). Cellulose synthases: new insights from crystallography and modeling. Trends Plant Sci. 19: 99–106. [DOI] [PubMed] [Google Scholar]
- Smith M.M., Stone B.A. (1973a). Studies on Lolium multiflorum endosperm in tissue culture I. Nutrition. Aust. J. Biol. Sci. 26: 123–134. [Google Scholar]
- Smith M.M., Stone B.A. (1973b). Chemical composition of the cell walls of Lolium multiflorum endosperm. Phytochemistry 12: 1361–1367. [Google Scholar]
- Søgaard C., Stenbæk A., Bernard S., Hadi M., Driouich A., Scheller H.V., Sakuragi Y. (2012). GO-PROMTO illuminates protein membrane topologies of glycan biosynthetic enzymes in the Golgi apparatus of living tissues. PLoS ONE 7: e31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos S.J., Fisher P.R., Stone B.A., Stanisich V.A. (1999). Detection of two loci involved in (1→3)-β-glucan (curdlan) biosynthesis by Agrobacterium sp. ATCC31749, and comparative sequence analysis of the putative curdlan synthase gene. Glycobiology 9: 31–41. [DOI] [PubMed] [Google Scholar]
- Suliman M., Chateigner-Boutin A.L., Francin-Allami M., Partier A., Bouchet B., Salse J., Pont C., Marion J., Rogniaux H., Tessier D., Guillon F., Larré C. (2013). Identification of glycosyltransferases involved in cell wall synthesis of wheat endosperm. J. Proteomics 78: 508–521. [DOI] [PubMed] [Google Scholar]
- Taketa S., Yuo T., Tonooka T., Tsumuraya Y., Inagaki Y., Haruyama N., Larroque O., Jobling S.A. (2012). Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-D-glucan biosynthesis. J. Exp. Bot. 63: 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonooka T., Aoki E., Yoshioka T., Taketa S. (2009). A novel mutant gene for (1-3, 1-4)-β-D-glucanless grain on barley (Hordeum vulgare L.) chromosome 7H. Breed. Sci. 59: 47–54. [Google Scholar]
- Trafford K., Haleux P., Henderson M., Parker M., Shirley N.J., Tucker M.R., Fincher G.B., Burton R.A. (2013). Grain development in Brachypodium and other grasses: possible interactions between cell expansion, starch deposition, and cell-wall synthesis. J. Exp. Bot. 64: 5033–5047. [DOI] [PubMed] [Google Scholar]
- Trethewey J.A.K., Harris P.J. (2002). Location of (1→3),(1→4)-β-D-glucans in vegetative cell walls of barley (Hordeum vulgare) using immunogold labelling. New Phytol. 154: 347–358. [DOI] [PubMed] [Google Scholar]
- Urbanowicz B.R., Rayon C., Carpita N.C. (2004). Topology of the maize mixed linkage (1->3),(1->4)-beta-d-glucan synthase at the Golgi membrane. Plant Physiol. 134: 758–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez M.E., et al. (2012). Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159: 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel P.H., DeAngelis P.L. (2007). Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J. Biol. Chem. 282: 36777–36781. [DOI] [PubMed] [Google Scholar]
- Widell S., Lundborg T., Larsson C. (1982). Plasma membranes from oats prepared by partition in an aqueous polymer two-phase system : on the use of light-induced cytochrome B reduction as a marker for the plasma membrane. Plant Physiol. 70: 1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.M., Bacic A. (2012). Preparation of plant cells for transmission electron microscopy to optimize immunogold labeling of carbohydrate and protein epitopes. Nat. Protoc. 7: 1716–1727. [DOI] [PubMed] [Google Scholar]
- Wilson S.M., Burton R.A., Doblin M.S., Stone B.A., Newbigin E.J., Fincher G.B., Bacic A. (2006). Temporal and spatial appearance of wall polysaccharides during cellularization of barley (Hordeum vulgare) endosperm. Planta 224: 655–667. [DOI] [PubMed] [Google Scholar]
- Xue J., Bosch M., Knox J.P. (2013). Heterogeneity and glycan masking of cell wall microstructures in the stems of Miscanthus x giganteus, and its parents M. sinensis and M. sacchariflorus. PLoS ONE 8: e82114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Johns M.A., Cao H., Rupani M. (2014). A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genomics 15: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.F., Staehelin L.A. (1992). Functional compartmentation of the Golgi apparatus of plant cells: immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol. 99: 1070–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- While this article was under review, an article by Kim et al. (2015) was published in which they heterologously expressed in tobacco yellow fluorescent protein (YFP) fused to the NH2-terminus of Brachypodium distachyon CSLF6. They demonstrated that the functional YFP fusion protein is localized to the Golgi apparatus and that the catalytic, NH2-, and COOH-termini are exposed in the cytosol, consistent with the findings reported here. Furthermore, their demonstration of a functional BdCSLF6 in yeast supports our hypothesis that CSLF6 is a bifunctional enzyme capable of both β-(1,3)- and β-(1,4)-transferase activity. [Google Scholar]
- Kim S.-J., Zemelis S., Keegstra K., Brandizzi F. (2015). The cytoplasmic localization of the catalytic site of CSLF6 supports a channeling model for the biosynthesis of mixed-linkage glucan. Plant J. 81: 537–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.